Finally, I got access to the LSJL gene expression data of genes upregulated over 2.0 fold. Based on information about Interstitial Fluid Flow, Dynamic Compression, and Hydrostatic Pressure stimulus we knew that LSJL could likely induce chondrogenesis because all of those stimuli have been associated with chondroinduction and LSJL likely induces all those stimuli but the LSJL chondroinduction was confirmed by the statistically significant increases in Sox9, COL2A1, and Agc1. The chondroinducer CCL2 was upregulated 3.691 fold. As a bonus, MATN3 was upregulated as well which is only expressed in cartilagenous tissues. In addition, HMGA2 and Lin28B were expressed which are associated with overgrowth. However, Growth Hormone expression decressed. GH levels went down by half and SOCS3 expression increased. The only thing missing was Sox5 & Sox6 expression but they could have been expressed below 2 fold. Also, H19 increased in expression which is associated with IGF2. LSJL also upregulates ITGBL1 which may be involved in mesenchymal condensation(Beta-1Integrin is involved in the integrin if ITGBL is like Beta-1integrin as suggested in the name it may induce condensation as well. According to Identification of two novel chromosome regions associated with isolated growth hormone deficiency., ITGBL1 homozygous deletion causes GH deficiency. ITGBL1 homozygous deletion also reduced height.
The genes were all analyzed for statistical significance and fold changes under 2 were not included thus there is low probability that the change expression is not statistically significant to LSJL.
The mouse did have open growth plates, they were female, and were 14 weeks of age. However, the genes were determined by bone samples in 4 mouse groups. The bones were entirely grounded.
In all 4 Control Samples, Sox9 expression was below 1. However, in 3 of the 4 Sox9 expression increased to between 2 and 4. In only one of the samples did expression decrease(Cluster 1). Similar for COL2A1, expression was below 1 for 1 through 4 and increased above 1 for 2 to 4. Same change for Agc1. However, for MATN3 Clusters 1,3,4 were below 1 and all increase above 1, however cluster 2 was above 1 and decreased below 1.
A level of 1 means that it's mRNA is synthesized greater than GADPH. A level less than 1 means it's synthesized less than GADPH. GADPH is a control molecule that is stably expressed in most tissues and cells at high levels. However, GADPH expression can alter in different cells in some conditions so that could have altered the data.
It's not likely that 3 out of 4 bone samples all contained growth plates indicating that Sox9 increased in expression in periosteum cells, Osteoblasts, Osteoclasts, fibroblasts, nerve cells, or Stem Cells.
According to Evolution of the osteoblast: skeletogenesis in gar and zebrafish., Sox9 can be expressed in osteoblasts. However, I haven't seen any data that states that Col2A1 or Agc1 can be expressed in osteoblast(or osteoclast) cells. Fibroblasts are the most likely cells that could be associated with those genes but there was still the increase in MATN3 expression which is solely associated with chondrogenesis.
So we have solid evidence that LSJL can induce new chondrogenesis.
LSJL upregulates several genes associated with neurons. Serotonin Receptors were upregulated 10 fold. There is very likely to be a conditioning mechanism. That is why I am now altering 4 days LSJL on legs/4 days LSJL on arms. Although gene expression continued to elevate on selected genes for 1 week but was almost eliminated on 2 weeks. Another one of CH Turners studies showed different genes started to be expressed after 4 days. I am now loading for 4 minutes.
Since now we have proven that LSJL can induce chondrogenesis in ectopic areas, lack of results could be either ineffective loading(a clamp is not a pizeoelectric mechanical loader) or due to adaptation which can be fixed by deconditioning periods? Although several people have performed irregular loading which would allow for decondition time.
Growing Taller: How Mesenchymal Stem Cells, Microfractures, Hydrostatic Pressure, and Periosteum makes increasing height possible
Thursday, August 30, 2012
Wednesday, August 29, 2012
Optimal Loads for Lateral Joint Loading
I finally got the full LSJL gene expression data and it shows that LSJL upregulates Sox9, COL2A1, Agc1, and MATN3 which is only expressed by cartilage all by at least 2 fold(See below). Decreased expression of Id2 is also associated with chondrogenesis. This provides support that LSJL can form new growth plates. I'll have to finish my analysis to ascertain more insight.
Recently, David stated that he had been loading his right epiphysis by 120lbs(!) to try to correct a 1cm length discrepancy. David used a hydraulic car jack and put two 45 lbs plates on his couch then he gradually used the jack to increase the pressure on his epiphysis. Here's his ankle picture after several months:
You can see his right epiphysis is much bigger than his left. David has reported no length change. An increase in bone size is excellent news because it indicates some of or some combination of the below: stem cell differentiation into osteoblasts which deposit new bone; mechanical signaling triggering existing osteoblasts to lay down new bone(bone modeling); and shear strain on periosteum resulting in periosteal progenitor cells which differentiate into osteoblasts which deposit new bone. Unfortunately, no height growth means no differentiation into chondroctyes. Could excessive loads favor osteogenesis over chondrogenesis? Or do you also need to load the articular cartilage to activate certain factors that favor chondrogenesis like Sox9 and TGF-Beta?
Signalling cascades in mechanotransduction: cell-matrix interactions and mechanical loading.
"Mechanical loading of articular cartilage stimulates the metabolism of resident chondrocytes and induces the synthesis of molecules to maintain the integrity of the cartilage[the articular cartilage connects to the hyaline cartilage of the growth plate so it can stimulate metabolism and synthesis of melcules there as well]. Mechanical signals modulate biochemical activity and changes in cell behavior through mechanotransduction. Compression of cartilage results in complex changes within the tissue including matrix and cell deformation, hydrostatic and osmotic pressure, fluid flow, altered matrix water content, ion concentration and fixed charge density[compression of cartilage changes hydrostatic pressure and one can presume growth plate hydrostatic pressure]. These changes are detected by mechanoreceptors on the cell surface, which include mechanosensitive ion channels and integrins that on activation initiate intracellular signalling cascades leading to tissue remodelling. Excessive mechanical loading also influences chondrocyte metabolism but unlike physiological stimulation leads to a quantitative imbalance between anabolic and catabolic activity resulting in depletion of matrix components[You shouldn't use too much load or risk depleting matrix components]. In this article we focus on the role of mechanical signalling in the maintenance of articular cartilage, and discuss how alterations in normal signalling can lead to pathology."
The bone begins as completely hyaline cartilage. Any mechanical loading induced growth should lead to increased long bone growth as the articular cartilage is connected to the resting zone hyaline cartilage until later in development.
"Increased joint loading in athletes is associated with an increase in the area of the load-bearing surface rather than an increase in cartilage thickness"<-Joint Loading increases joint width but not height but can lateral loading of joints encourage joint height as it's on a different axis?
"cyclic tensile loading increased the mRNA level of MMP-1, MMP-3, MMP-9, IL-1β, TNF-α and TIMP-1 in cultured chondrocytes"<-tensile loading means stretch. Aside from Interleukin Beta and TNF-Alpha these are the best genes for height growth to be expressed by chondrocytes. So lateral joint loading directly on the cartilage may have height increase benefits as well.
"insulin-like growth factor-1 (IGF-1) and TGF-β increase chondrocyte cell surface expression of α3/α5 integrin subunits and stimulate adhesion of chondrocytes to fibronectin and type II collagen"<-IGF-1 and TGF-Beta encourage chondrocytes to adhere to the Type II collagen parts of the growth plate
"IL-4 not only mediates anabolic signalling by increasing aggrecan expression but can decrease catabolic events. In an animal model intra-articular injection of IL-4 decreased chondrocyte nitric oxide production and inhibited destruction of cartilage in instability-induced osteoarthritis. Pre-treatment with IL-4 (10 ng/mL) suppressed both MMP-13 and cathepsin B induction by mechanical stress, as well as cyclical tensile stress-induced IL-1β expression"<-IL-4 may be a promising height increase supplement for the future.
Gene expression profiles of dynamically compressed single chondrocytes and chondrons.
"A chondrocyte produces a hydrated pericellular matrix (PCM); together they form a chondron. Previous work has shown that the presence of the PCM influences the biological response of chondrocytes to loading. The objective of this study was to determine the gene expression profiles of enzymatically isolated single chondrocytes and chondrons in response to dynamic compression. Cartilage specific extracellular matrix components and transcription factors were examined. Following dynamic compression, chondrocytes and chondrons showed variations in gene expression profiles. Aggrecan[increases proteoglycan content], Type II collagen[Type II collagen is the basis for articular and hyaline cartilage, could possibly lead to height growth] and osteopontin[involved in bone modeling] gene expression were significantly increased in chondrons. Lubricin gene expression decreased[Lubricin is a joint lubricant so could be part of a negative feedback mechanism] in both chondrons and chondrocytes. Dynamic compression had no effect on SOX9 gene expression. Our results demonstrate a clear role for the PCM in interfacing the mechanical signalling in chondrocytes in response to dynamic compression. Further investigation of single chondrocytes and chondrons from different zones within articular cartilage may further our understanding of cartilage mechanobiology."
Dynamic loading of articular cartilage may enhance height growth by upregulating Type II Collagen expression.
[Type II collagen fragment capacity to induce collagen cleavage and hypertrophy of articular chondrocyte]
"The objective of this study was to determine whether a peptide of type II collagen which can induce collagenase activity can also induce chondrocyte differentiation (hypertrophy) in articular cartilage. At high but naturally occurring concentrations (10 microM and up) the collagen peptide CB12-II induced an increase in the expressions of MMP-13 (24h) and cleavage of type II collagen by collagenase in the mid zone (day 4) and also in the superficial zone (day 6). Furthermore the peptide induced an increase in proliferation on day 1 in the mid and deep zones extending to the superficial zone by day 4. There was also upregulation of COL10A1 expression at day 4 and of type X staining in the mid zone extending to the superficial zone by day 6. Apoptopic cell death was increased by day 4 in the lower deep zone and also in the superficial zone at day 7. The increase in apoptosis in the deep zone was also seen in controls. Our results show that the induction of collagenase activity by cryptic peptide sequence of type II collagen is accompanied by chondrocyte hypertrophy and associated cellular and matrix changes. This induction occurs in the mid and superficial zones of previously healthy articular cartilage. This response of the chondrocyte to a cryptic sequence of denaturated type II collagen may play a role in naturally occurring hypertrophy in endochondral ossification and in the development of cartilage pathology in osteoarthritis."
So the Type II Collagen environment in the growth plate can trigger chondrocyte hypertrophy. This could be why proliferative capacity is conserved in growth hormone deficiency as there is less surrounding Type II Collagen. So overloading may result in the denaturing of Type II collagen which encourages ossification.
Genome-Wide Analyses of Gene Expression during Mouse Endochondral Ossification
"Numerous molecular markers characterize the central stages of the chondrocyte life cycle. Chondrogenesis is typified by the expression of Sox transcription factors 5,6 and 9. Proliferating chondrocytes synthesize an ECM composed mainly of collagen II and aggrecan, among others, while the central ECM molecule expressed in hypertrophic cartilage is collagen X. Factors expressed at the chondro-osseous junction regulate chondrocyte apoptosis and mineralization of the cartilaginous ECM. Late hypertrophic chondrocytes express factors that promote angiogenesis, bone deposition and the secretion of bone-specific cell ECM[So chondrocyte hypertrophy encourages ossification without estrogen]. These factors include Vegf (vascular endothelial growth factor), Mmp13(matrix metalloproteinase 13), Mmp9 and Ibsp. Additional markers of the osteoblast and osteoclast phenotype, including core-binding factor alpha 1/runt-related transcription factor 2 (Cbfa1/Runx2), acid phosphatase 5, tartrate resistant (Acp5) and tumor necrosis factor (ligand) superfamily, member 11 (Tnfsf11; RANKL/receptor activator of NF-kappaB ligand) are upregulated in hypertrophic cartilage and cells in the zone of ossification."
"When chondrocytes terminally differentiate, they undergo apoptosis, leaving behind a calcified extracellular matrix (ECM) that is remodeled and degraded by invading blood vessels, osteoprogenitor cells and bone-resorbing cells."<-terminal differentiation proceeds fusion which involves remodeling left over Extracellular Matrix. Ossification does not directly oppose cartilage growth.
"the expression of early stage chondrocyte markers such as Sox family members 5,6 and 9 (Sox5,6 and 9), Col2a1, growth differentiation factor 5 (Gdf5), Agc 1, Col11a1, Hapln1, Fgfr3 and Col9a2"<-These are the genes we are looking to be expressed by LSJL, not necessarily immediately but they should be expressed after LSJL is performed to indicate that LSJL was effective at inducing a new growth plate.
"several known growth plate markers [include] Sox9, Col2a1, Ihh and Cdkn1c"
Here's the list of genes upregulated by LSJL:
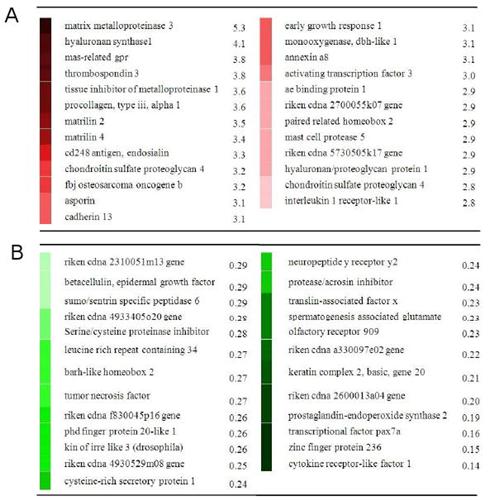
Column A are upregulated genes whereas Column B are downregulated. The genes involved are involved in the PI3K pathway and TGF-Beta pathway both of which are important to height growth. Perhaps, loading the cartilage is important to activate these genes and pathways. Note that the genes expressed shouldn't match the growth plate genes right away as it likely takes time for the growth plates to be formed. However, you should expect to see growth plate genes eventually.
Here's additional genes modified by LSJL:
Under expressed proteins are blue and overexpressed are red. There are no underexpressed proteins in the two pathways. When one point shares more proteins for example BMP's some proteins can be underexpressed and some over.
Other genes of note:
" Genes highlighted in those pathways include inositol 1,4,5-triphosphate 3-kinase and phospholipase C (plc) in PI3K, collagen (col3α1, col4α1, col6α1), integrin β4, and thrombospondin 3 in ECM-receptor interaction, TGFβ receptor 1 and smad1in TGFβ signaling pathway, and wnt2, frizzled 2 and Wnt1-inducible protein 2 in Wnt signaling pathway."
Single genes that can be identified as being overexpressed: Integrin Alpha V(CD51) and Integrin Alpha 11.
Genes that were upregulated above 2 fold or below .5 fold compared to control samples(Bolded genes are pro-chondrogenic):
Barx2
CDC42bpa
Zfp9
Cdh10
FGF2-2.433
ECM2-2.027
Dmrt2
Zfp533-2.188(upregulated during cartilage formation)
Gli3-3.184(one form of Gli3 may inhibit chondrocyte proliferation)
Fzd3
Nov-3.563
THBS2
Aspn
Dkk3
Itgbl1
Scx
H19-2.174(H19 expression is liked to IGF-2)
Grem2
FGFR1
Cav3-2.001(Myostatin inhibitor)
MMP2
THBS4
WISP2
SOCS3-2.484
TUBB6
Sox9-3.148
IGFBP6-2.083
Pax1-2.166
Agc1-2.474
Jun
Zfp36
Dnm1-2.352
Gas1
HMGA2-2.137(increased expression can cause overgrowth)
Dhh
COL2A1-2.012
MATN4-2.166
Zfp410
IRS1-0.339
PLRG1
TRAPPC3
TLR7
NPC1
Zfp148
Zfp106
HMGB2-0.291(reduction in HMGB2 is associated with apoptosis, according to Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation., HMGB2 inhibits chondrogenesis)
Zfp313
GPR108
Kif22
Zfp46
Rad17
Smad1-0.48
MAPK6
ADAMTS7-0.443
Zfp692
Zfp75
Zfp27
MAPK8
Zfp238
Id2-0.459(associated with early chondrogenic aggregates)
Gad1
GH-0.498
Lin9-0.471
STAT 5B-0.424(increases GH signaling)
Vcam 1-0.487(expressed by bone marrow stem cells, decrease in expression could indicate an increase in differentiation)
Accn1
MATN2-2.735
Recently, David stated that he had been loading his right epiphysis by 120lbs(!) to try to correct a 1cm length discrepancy. David used a hydraulic car jack and put two 45 lbs plates on his couch then he gradually used the jack to increase the pressure on his epiphysis. Here's his ankle picture after several months:

You can see his right epiphysis is much bigger than his left. David has reported no length change. An increase in bone size is excellent news because it indicates some of or some combination of the below: stem cell differentiation into osteoblasts which deposit new bone; mechanical signaling triggering existing osteoblasts to lay down new bone(bone modeling); and shear strain on periosteum resulting in periosteal progenitor cells which differentiate into osteoblasts which deposit new bone. Unfortunately, no height growth means no differentiation into chondroctyes. Could excessive loads favor osteogenesis over chondrogenesis? Or do you also need to load the articular cartilage to activate certain factors that favor chondrogenesis like Sox9 and TGF-Beta?
Signalling cascades in mechanotransduction: cell-matrix interactions and mechanical loading.
"Mechanical loading of articular cartilage stimulates the metabolism of resident chondrocytes and induces the synthesis of molecules to maintain the integrity of the cartilage[the articular cartilage connects to the hyaline cartilage of the growth plate so it can stimulate metabolism and synthesis of melcules there as well]. Mechanical signals modulate biochemical activity and changes in cell behavior through mechanotransduction. Compression of cartilage results in complex changes within the tissue including matrix and cell deformation, hydrostatic and osmotic pressure, fluid flow, altered matrix water content, ion concentration and fixed charge density[compression of cartilage changes hydrostatic pressure and one can presume growth plate hydrostatic pressure]. These changes are detected by mechanoreceptors on the cell surface, which include mechanosensitive ion channels and integrins that on activation initiate intracellular signalling cascades leading to tissue remodelling. Excessive mechanical loading also influences chondrocyte metabolism but unlike physiological stimulation leads to a quantitative imbalance between anabolic and catabolic activity resulting in depletion of matrix components[You shouldn't use too much load or risk depleting matrix components]. In this article we focus on the role of mechanical signalling in the maintenance of articular cartilage, and discuss how alterations in normal signalling can lead to pathology."
The bone begins as completely hyaline cartilage. Any mechanical loading induced growth should lead to increased long bone growth as the articular cartilage is connected to the resting zone hyaline cartilage until later in development.
"Increased joint loading in athletes is associated with an increase in the area of the load-bearing surface rather than an increase in cartilage thickness"<-Joint Loading increases joint width but not height but can lateral loading of joints encourage joint height as it's on a different axis?
"cyclic tensile loading increased the mRNA level of MMP-1, MMP-3, MMP-9, IL-1β, TNF-α and TIMP-1 in cultured chondrocytes"<-tensile loading means stretch. Aside from Interleukin Beta and TNF-Alpha these are the best genes for height growth to be expressed by chondrocytes. So lateral joint loading directly on the cartilage may have height increase benefits as well.
"insulin-like growth factor-1 (IGF-1) and TGF-β increase chondrocyte cell surface expression of α3/α5 integrin subunits and stimulate adhesion of chondrocytes to fibronectin and type II collagen"<-IGF-1 and TGF-Beta encourage chondrocytes to adhere to the Type II collagen parts of the growth plate
"IL-4 not only mediates anabolic signalling by increasing aggrecan expression but can decrease catabolic events. In an animal model intra-articular injection of IL-4 decreased chondrocyte nitric oxide production and inhibited destruction of cartilage in instability-induced osteoarthritis. Pre-treatment with IL-4 (10 ng/mL) suppressed both MMP-13 and cathepsin B induction by mechanical stress, as well as cyclical tensile stress-induced IL-1β expression"<-IL-4 may be a promising height increase supplement for the future.
Gene expression profiles of dynamically compressed single chondrocytes and chondrons.
"A chondrocyte produces a hydrated pericellular matrix (PCM); together they form a chondron. Previous work has shown that the presence of the PCM influences the biological response of chondrocytes to loading. The objective of this study was to determine the gene expression profiles of enzymatically isolated single chondrocytes and chondrons in response to dynamic compression. Cartilage specific extracellular matrix components and transcription factors were examined. Following dynamic compression, chondrocytes and chondrons showed variations in gene expression profiles. Aggrecan[increases proteoglycan content], Type II collagen[Type II collagen is the basis for articular and hyaline cartilage, could possibly lead to height growth] and osteopontin[involved in bone modeling] gene expression were significantly increased in chondrons. Lubricin gene expression decreased[Lubricin is a joint lubricant so could be part of a negative feedback mechanism] in both chondrons and chondrocytes. Dynamic compression had no effect on SOX9 gene expression. Our results demonstrate a clear role for the PCM in interfacing the mechanical signalling in chondrocytes in response to dynamic compression. Further investigation of single chondrocytes and chondrons from different zones within articular cartilage may further our understanding of cartilage mechanobiology."
Dynamic loading of articular cartilage may enhance height growth by upregulating Type II Collagen expression.
[Type II collagen fragment capacity to induce collagen cleavage and hypertrophy of articular chondrocyte]
"The objective of this study was to determine whether a peptide of type II collagen which can induce collagenase activity can also induce chondrocyte differentiation (hypertrophy) in articular cartilage. At high but naturally occurring concentrations (10 microM and up) the collagen peptide CB12-II induced an increase in the expressions of MMP-13 (24h) and cleavage of type II collagen by collagenase in the mid zone (day 4) and also in the superficial zone (day 6). Furthermore the peptide induced an increase in proliferation on day 1 in the mid and deep zones extending to the superficial zone by day 4. There was also upregulation of COL10A1 expression at day 4 and of type X staining in the mid zone extending to the superficial zone by day 6. Apoptopic cell death was increased by day 4 in the lower deep zone and also in the superficial zone at day 7. The increase in apoptosis in the deep zone was also seen in controls. Our results show that the induction of collagenase activity by cryptic peptide sequence of type II collagen is accompanied by chondrocyte hypertrophy and associated cellular and matrix changes. This induction occurs in the mid and superficial zones of previously healthy articular cartilage. This response of the chondrocyte to a cryptic sequence of denaturated type II collagen may play a role in naturally occurring hypertrophy in endochondral ossification and in the development of cartilage pathology in osteoarthritis."
So the Type II Collagen environment in the growth plate can trigger chondrocyte hypertrophy. This could be why proliferative capacity is conserved in growth hormone deficiency as there is less surrounding Type II Collagen. So overloading may result in the denaturing of Type II collagen which encourages ossification.
Genome-Wide Analyses of Gene Expression during Mouse Endochondral Ossification
"Numerous molecular markers characterize the central stages of the chondrocyte life cycle. Chondrogenesis is typified by the expression of Sox transcription factors 5,6 and 9. Proliferating chondrocytes synthesize an ECM composed mainly of collagen II and aggrecan, among others, while the central ECM molecule expressed in hypertrophic cartilage is collagen X. Factors expressed at the chondro-osseous junction regulate chondrocyte apoptosis and mineralization of the cartilaginous ECM. Late hypertrophic chondrocytes express factors that promote angiogenesis, bone deposition and the secretion of bone-specific cell ECM[So chondrocyte hypertrophy encourages ossification without estrogen]. These factors include Vegf (vascular endothelial growth factor), Mmp13(matrix metalloproteinase 13), Mmp9 and Ibsp. Additional markers of the osteoblast and osteoclast phenotype, including core-binding factor alpha 1/runt-related transcription factor 2 (Cbfa1/Runx2), acid phosphatase 5, tartrate resistant (Acp5) and tumor necrosis factor (ligand) superfamily, member 11 (Tnfsf11; RANKL/receptor activator of NF-kappaB ligand) are upregulated in hypertrophic cartilage and cells in the zone of ossification."
"When chondrocytes terminally differentiate, they undergo apoptosis, leaving behind a calcified extracellular matrix (ECM) that is remodeled and degraded by invading blood vessels, osteoprogenitor cells and bone-resorbing cells."<-terminal differentiation proceeds fusion which involves remodeling left over Extracellular Matrix. Ossification does not directly oppose cartilage growth.
"the expression of early stage chondrocyte markers such as Sox family members 5,6 and 9 (Sox5,6 and 9), Col2a1, growth differentiation factor 5 (Gdf5), Agc 1, Col11a1, Hapln1, Fgfr3 and Col9a2"<-These are the genes we are looking to be expressed by LSJL, not necessarily immediately but they should be expressed after LSJL is performed to indicate that LSJL was effective at inducing a new growth plate.
"several known growth plate markers [include] Sox9, Col2a1, Ihh and Cdkn1c"
Here's the list of genes upregulated by LSJL:

Column A are upregulated genes whereas Column B are downregulated. The genes involved are involved in the PI3K pathway and TGF-Beta pathway both of which are important to height growth. Perhaps, loading the cartilage is important to activate these genes and pathways. Note that the genes expressed shouldn't match the growth plate genes right away as it likely takes time for the growth plates to be formed. However, you should expect to see growth plate genes eventually.
Here's additional genes modified by LSJL:
Under expressed proteins are blue and overexpressed are red. There are no underexpressed proteins in the two pathways. When one point shares more proteins for example BMP's some proteins can be underexpressed and some over.
Other genes of note:
" Genes highlighted in those pathways include inositol 1,4,5-triphosphate 3-kinase and phospholipase C (plc) in PI3K, collagen (col3α1, col4α1, col6α1), integrin β4, and thrombospondin 3 in ECM-receptor interaction, TGFβ receptor 1 and smad1in TGFβ signaling pathway, and wnt2, frizzled 2 and Wnt1-inducible protein 2 in Wnt signaling pathway."
Single genes that can be identified as being overexpressed: Integrin Alpha V(CD51) and Integrin Alpha 11.
Genes that were upregulated above 2 fold or below .5 fold compared to control samples(Bolded genes are pro-chondrogenic):
Barx2
CDC42bpa
Zfp9
Cdh10
FGF2-2.433
ECM2-2.027
Dmrt2
Zfp533-2.188(upregulated during cartilage formation)
Gli3-3.184(one form of Gli3 may inhibit chondrocyte proliferation)
Fzd3
Nov-3.563
THBS2
Aspn
Dkk3
Itgbl1
Scx
H19-2.174(H19 expression is liked to IGF-2)
Grem2
FGFR1
Cav3-2.001(Myostatin inhibitor)
MMP2
THBS4
WISP2
SOCS3-2.484
TUBB6
Sox9-3.148
IGFBP6-2.083
Pax1-2.166
Agc1-2.474
Jun
Zfp36
Dnm1-2.352
Gas1
HMGA2-2.137(increased expression can cause overgrowth)
Dhh
COL2A1-2.012
MATN4-2.166
Zfp410
IRS1-0.339
PLRG1
TRAPPC3
TLR7
NPC1
Zfp148
Zfp106
HMGB2-0.291(reduction in HMGB2 is associated with apoptosis, according to Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation., HMGB2 inhibits chondrogenesis)
Zfp313
GPR108
Kif22
Zfp46
Rad17
Smad1-0.48
MAPK6
ADAMTS7-0.443
Zfp692
Zfp75
Zfp27
MAPK8
Zfp238
Id2-0.459(associated with early chondrogenic aggregates)
Gad1
GH-0.498
Lin9-0.471
STAT 5B-0.424(increases GH signaling)
Vcam 1-0.487(expressed by bone marrow stem cells, decrease in expression could indicate an increase in differentiation)
Accn1
MATN2-2.735
MATN3-2.485(Matrilin 3 is specific to cartilage matrix proteins)
Nkx3.2(also known as Bpax1)-0.49(this gene is pro-chondrogenic so it may be downregulated as part of a negative feedback mechanism)
Wnt2-3.089
Hey2-2.685
ADAMTS1-2.674
Hes1-2.405
Smad9-2.383
dusp14-2.32
Arg1-2.25
Sulf1-2.195
Vcan-2.144
Lin28B-2.143(Increases HMGA2 activity which in turn should increase height)
MMP14-2.055
HHIP-2.014
MMP7-0.491
CAMK2G-0.458
TGFBR1-0.411
Arg2-0.36
HTR2C-10.47(Serotonin Receptor)
PTGS2-7.63(synthesizes prostoglandin)
TNMD-6.185(Tendon Molecular marker)
IL6-3.051(activates STAT3)
BMPR1B- 2.166
COL9A1-2.455
COL11A1-2.029
HAPLN1-2.585(expressed in early chondrogenesis, associated with hyaluronic acid binding)
Dpt-3.144(increases in expression throughout chondrogenesis)
PRKG2-2.894(associated with cGMP)
Ptn-2.649(regulates cell cycle)
PDGFC-2.1
PKIA-2.013(inhibits cAMP)
CCNB1-0.437(cell cycle progression)
Sdc2-0.426(Syndecan 2, involved in pre-chondrogenic differentiation)
PDE6H-0.424(associated with cAMP)
HABP4-0.417(binds with hyaluronic acid)
CREB3L1-2.238(also called OASIS, may increase GH and IGF-1 levels)
COL10A1-2.012
HTRA1-2.322(interacts with BMP4, Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. states that HtrA1 is involved in Aggrecan breakdown)
HES5-2.562(In the data as BHLHB5)
GNAS-0.329(implicated in some forms of heterotropic ossification)
According to another LSJL study LSJL suppresses Sost and Dkk1.
Here's how these genes compare to MSCs normally undergoing endochondral ossification.
Gene Expression Profile during Chondrogenesis in Human Bone Marrow derived Mesenchymal Stem Cells using a cDNA Microarray.
"Chondrogenesis was induced by culturing human bone marrow (BM) derived MSCs in micromass pellets in the presence of defined medium for 3, 7, 14 or 21 days. Several genes regulated during chondrogenesis were then identified by reverse transcriptase-polymerase chain reaction (RT-PCR). Using an ABI microarray system, we determined the differential gene expression profiles of differentiated chondrocytes and BM-MSCs. Normalization of this data resulted in the identification of 1,486 differentially expressed genes. To verify gene expression profiles determined by microarray analysis, the expression levels of 10 genes with high fold changes were confirmed by RT-PCR. Gene expression patterns of 9 genes (Hrad6B, annexinA2, BMP-7, contactin-1, peroxiredoxin-1, heat shock transcription factor-2, synaptotagmin IV, serotonin receptor-7, Axl) in RT-PCR were similar to the microarray gene expression patterns."
Note that the LSJL gene expression study was done on rats whereas this was done on humans. It's also possible that these genes were expressed at lower levels than were noted in the study. Note there is definite signs of chondrogenesis like the genes related to proteoglycans.
These are different than the highly expressed genes in the LSJL study, AnnexinA2 facilitates actin cytoskeleton formation whereas in the LSJL study AnnexinA8 was present. AnnexinA8 is an anticoagulant so it stops blood clotting likely to reduce hydrostatic pressure.
"The antigen which bound to SH2 antibody was identified as endoglin (CD105), a receptor for TGF-β3, which potentially plays a role in mediating the chondrogenic differentiation of MSCs and in their interactions with hematopoietic cells"<-endosialin is present instead in LSJL. Endosialin relates to calcium ion binding.
"Axl, synaptotagmin IV, Hrad6B, peroxiredoxin-1, BMP-7, heat shock transcription factor-2, annexin A2, contactin-1 and serotonin receptor-7 expressions were maintained in differentiating BM-MSCs until day 14."<-So maybe upregulating serotonin expression can upregulate chondrogenesis?
Wnt2-3.089
Hey2-2.685
ADAMTS1-2.674
Hes1-2.405
Smad9-2.383
dusp14-2.32
Arg1-2.25
Sulf1-2.195
Vcan-2.144
Lin28B-2.143(Increases HMGA2 activity which in turn should increase height)
MMP14-2.055
HHIP-2.014
MMP7-0.491
CAMK2G-0.458
TGFBR1-0.411
Arg2-0.36
HTR2C-10.47(Serotonin Receptor)
PTGS2-7.63(synthesizes prostoglandin)
TNMD-6.185(Tendon Molecular marker)
IL6-3.051(activates STAT3)
BMPR1B- 2.166
COL9A1-2.455
COL11A1-2.029
HAPLN1-2.585(expressed in early chondrogenesis, associated with hyaluronic acid binding)
Dpt-3.144(increases in expression throughout chondrogenesis)
PRKG2-2.894(associated with cGMP)
Ptn-2.649(regulates cell cycle)
PDGFC-2.1
PKIA-2.013(inhibits cAMP)
CCNB1-0.437(cell cycle progression)
Sdc2-0.426(Syndecan 2, involved in pre-chondrogenic differentiation)
PDE6H-0.424(associated with cAMP)
HABP4-0.417(binds with hyaluronic acid)
CREB3L1-2.238(also called OASIS, may increase GH and IGF-1 levels)
COL10A1-2.012
HTRA1-2.322(interacts with BMP4, Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. states that HtrA1 is involved in Aggrecan breakdown)
HES5-2.562(In the data as BHLHB5)
GNAS-0.329(implicated in some forms of heterotropic ossification)
According to another LSJL study LSJL suppresses Sost and Dkk1.
Here's how these genes compare to MSCs normally undergoing endochondral ossification.
Gene Expression Profile during Chondrogenesis in Human Bone Marrow derived Mesenchymal Stem Cells using a cDNA Microarray.
"Chondrogenesis was induced by culturing human bone marrow (BM) derived MSCs in micromass pellets in the presence of defined medium for 3, 7, 14 or 21 days. Several genes regulated during chondrogenesis were then identified by reverse transcriptase-polymerase chain reaction (RT-PCR). Using an ABI microarray system, we determined the differential gene expression profiles of differentiated chondrocytes and BM-MSCs. Normalization of this data resulted in the identification of 1,486 differentially expressed genes. To verify gene expression profiles determined by microarray analysis, the expression levels of 10 genes with high fold changes were confirmed by RT-PCR. Gene expression patterns of 9 genes (Hrad6B, annexinA2, BMP-7, contactin-1, peroxiredoxin-1, heat shock transcription factor-2, synaptotagmin IV, serotonin receptor-7, Axl) in RT-PCR were similar to the microarray gene expression patterns."
Note that the LSJL gene expression study was done on rats whereas this was done on humans. It's also possible that these genes were expressed at lower levels than were noted in the study. Note there is definite signs of chondrogenesis like the genes related to proteoglycans.
These are different than the highly expressed genes in the LSJL study, AnnexinA2 facilitates actin cytoskeleton formation whereas in the LSJL study AnnexinA8 was present. AnnexinA8 is an anticoagulant so it stops blood clotting likely to reduce hydrostatic pressure.
"Axl, synaptotagmin IV, Hrad6B, peroxiredoxin-1, BMP-7, heat shock transcription factor-2, annexin A2, contactin-1 and serotonin receptor-7 expressions were maintained in differentiating BM-MSCs until day 14."<-So maybe upregulating serotonin expression can upregulate chondrogenesis?
"BMP-7 is a strong chemotactic component in cartilage cells produced by mesenchymal stem cells, and it can promote cartilage cells to secrete specific extracellular matrix (proteoglycans and collagen type II). And BMP-7 can induce the differentiation of BM-MSCs into cartilage cells."
"Annexins bind to negatively charged phospholipids in a Ca2+-dependent manner."
"Contactin-1 is a cell surface adhesion molecule"
"IL-15 [which is expressed by MSCs] is a potent apoptosis inhibitor "
Hiroki Yokota wrote a paper that mentions gene expression in chondrogenesis:
Modelling and identification of transcription-factor binding motifs in human chondrogenesis.
Hiroki Yokota wrote a paper that mentions gene expression in chondrogenesis:
Modelling and identification of transcription-factor binding motifs in human chondrogenesis.
Red are downregulated genes and green are upregulated genes. Black is neutral. We're looking for genes upregulated 1 day as that would be the time frame closest to that of the LSJL gene expression study. LSJL genes were taken at 49 hours. Column A is the observed profile. ILR1(although in LSJL it's interleukin 1 receptor-like 1) and BMP2 is shared between chondrogenesis and LSJL.
The predicted regulatory model for Col2A1 "[predicts] that AP-1 and Smad would be the continuous stimulator, and Sox9 would be the inhibitor at day 1 and the stimulator at days 7 to 21". c-Fos which LSJL upregulates is part of the AP-1 complex.
"AP-1 is reported to play the critical role in differentiation as a target of chondrogenetic growth factor such as bone morphogenetic protein-2 (BMP-2)"
"The model predicted that AP-1 would be the strong stimulatory factor during chondrogenesis. NFkB is known to control expression of BMP-2 and Sox-9 genes"
"The Sox-9 gene is one of the essential transcription factors in chondrogenesis by activating the enhancer element of a series of chondrogenetic marker genes such as Col2a1; Col9a2; Col11a2; and Aggrecan"
Labels:
lateral synovial joint loading
Tuesday, August 28, 2012
The genes associated with human height development
Target genes for height increase are as follows, for KO genes supplements or activities that inhibit these genes will increase height. For OE genes supplements or activities that upregulate these genes will increase height. For biphasic genes both overexpression and knockout decrease height.
KO(inhibit these):
FANCC
NOG
NPR3
RNF135
SOCS2
STC2
GPC3
IGF2R
GPR30
POMC
FGF21
FGFR3
FGFR1
ICR1
OE(stimulate these):
NPPC
PLAG1
SHOX2
Twist1
CNP
IGF2
IGF1
Akt1
CTGF(CCN2)
Mef2c[chondrocyte specific]
Biphasic Genes(genes need optimal quality):
HHIP
Zfp521
LCN2
Although, the genes involved in juvenile growth are not the same as the ones involved with reinvigorating growth. Analyzing the genes involved in juvenile growth can help illuminate the pathways involved in endochondral ossification like the HMGA2 gene which is associated with chromatin and let-7.
Identification of ten loci associated with height highlights new biological pathways in human growth.
"Ten newly identified and two previously reported loci were strongly associated with variation in height (P values from 4 × 10-7 to 8 × 10-22). Together, these 12 loci account for ~2% of the population variation in height. Individuals with ≤8 height-increasing alleles and ≥16 height-increasing alleles differ in height by ~3.5 cm. The newly identified loci, along with several additional loci with strongly suggestive associations, encompass both strong biological candidates and unexpected genes, and highlight several pathways (let-7 targets, chromatin remodeling proteins and Hedgehog signaling) as important regulators of human stature. These results expand the picture of the biological regulation of human height and of the genetic architecture of this classical complex trait."<-We've looked at IHH and chromatin remodeling. Let-7 is associated with osteoblast and type I collagen which means that Type I Collagen and osteoblasts may be more important than first thought.
The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature.
"Human height is considered highly heritable and correlated with certain disorders, such as type 2 diabetes and cancer. Despite environmental influences, genetic factors are known to play an important role in stature determination. A number of genetic determinants of adult height have already been established through genome wide association studies.
To examine 51 single nucleotide polymorphisms (SNPs) corresponding to the 46 previously reported genomic loci for height in 8,184 European American children with height measurements. We leveraged genotyping data from our ongoing GWA study of height variation in children in order to query the 51 SNPs in this pediatric cohort.
Sixteen of these SNPs yielded at least nominally significant association to height, representing fifteen different loci including EFEMP1-PNPT1, GPR126, C6orf173, SPAG17, Histone class 1, HLA class III and GDF5-UQCC. Other loci revealed no evidence for association, including HMGA1 and HMGA2. For the 16 associated variants, the genotype score explained 1.64% of the total variation for height z-score.
Among 46 loci that have been reported to associate with adult height to date, at least 15 also contribute to the determination of height in childhood."
Genome-wide association analysis identifies 20 loci that influence adult height.
"Adult height is a model polygenic trait, but there has been limited success in identifying the genes underlying its normal variation. To identify genetic variants influencing adult human height, we used genome-wide association data from 13,665 individuals and genotyped 39 variants in an additional 16,482 samples. We identified 20 variants associated with adult height (P < 5 × 10−7, with 10 reachingP < 1 × 10−10). Combined, the 20 SNPs explain ~3% of height variation, with a ~5 cm difference between the 6.2% of people with 17 or fewer ‘tall’ alleles compared to the 5.5% with 27 or more ‘tall’ alleles. The loci we identified implicate genes in Hedgehog signaling (IHH, HHIP, PTCH1), extracellular matrix (EFEMP1, ADAMTSL3, ACAN) and cancer (CDK6, HMGA2, DLEU7) pathways, and provide new insights into human growth and developmental processes. Finally, our results provide insights into the genetic architecture of a classic quantitative trait."
List
-SNP in the 3′ UTR of the HMGA2 gene(Chromatin, Type I Collagen)(knockout causes growth defects)
-GDF5-UQCC(Chondrogenesis, may induce ectopic chondrogenesis thus could potentially be chondroinductive, KO reduces height, allele that increases height also reduces risk for osteoarthritis, affects peak height velocity during infancy)
-SH3GL3-ADAMTSL3( glycoprotein metalloprotease, regulates venous invasion)
-CDK6(associated with let-7, allele that associates with greater height also associates with greater risk of rheumatoid arthritis)
-CHCHD7-RDHE2
-ZBTB38(methyl-DNA-binding transcriptional repressor, methylation affects cellular senesence)
-GPR126(involved in myelination which is involved in nervous function thus nerves may be very important for height growth)
-HIST1H1D(compaction of chromatin into higher end structures, affects peak height velocity during infancy)
-HHIP(reduces Ihh signaling, HHIP overexpression reduces height, mouse knockout causes skeletal defects, affects peak height velocity during infancy)
-TRIP11-ATXN3(Thyroid Hormone Receptor, thyroid hormone is associated with IGF-1 receptors, mutations cause dwarfism)
-LIN28A
-Six6
-Sox5(involved in dwarfism)
-CDKAL1(affects birth length and weight)
-ADRB1(affects birth length and weight)
-LIN28B(associated with let-7)
-DOT1L(histone methyltransferase; histone modifies Sox9 transcription; associated with let-7, affects peak height velocity during puberty)
-PTCH1(SHH & IHH Receptor, knockout causes skeletal defects)
-ACAN(Aggrecan an ECM protein, KO reduces height)
-ADAMTS17(processes proteoglycans and cleaves ECM)
-ADAMTSL3(ECM-related, lacks metalloprotease and other distegrin-like domains unlike normal ADAMTS)
-SCMH1(maintains that transcriptively repressive state of some genes, modifies histone, mouse knockout causes skeletal defects)
-ANAPC13(controls progression through mitosis and G1 phase of the cell cycle)
-PRKG2(encodes the cGMP-dependent protein kinase II (cGKII); cGMP is associated with CNP and is involved in chondrocyte hypertrophy, upregulated in LSJL)
-NCAPG(required for conversion of interphase chromatin, essential for progression in mitosis)
-EFEMP1-PNPT1(EFEMP1 may play a role in cell adhesion and migration and may inhibit chondrocyte differentiation. EFEMP1 is also an ECM stabilizer. overexpression stimulates proliferation and supresses chondrocyte differentiation, knockout reduces body size)
-PLAG1(upregulates IGF-II, knockout causes growth defects, has a major effect on stature according to Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature.)
-SPAG17[upregulated by LSJL](maintains structural integrity of the sperm, may directly cause height or just be a correlation as it's important for parents to have SPAG17 but not yourself to maximize height)
-DLEU7(affects peak height velocity during infancy)
-Prkca
-SPAG1
-Slit3
-MOS
-Sox8(shortens rib length)
-Lyn
-CTNNB1
-AXIN2(Tgfb and Wnt involvement)
-Wnt4
-CHSY1(involved in TGFBeta signaling)
-FNDC3B(some diseases with decreased bone length have this involved)
-TRIOBP(involved with Beta-Catenin binding)
-BNC2(regulates cartilage differentiation, some mutations shorten ulna)
-WWP2(cartilage and chondrocyte development, affects PDGF signaling)
-Sp3(growth plate receptor signalling pathway)
-PLAGL1(mutations reduce height)
-Glt25d2
-C3orf47
-MTMR11
-C3orf63
-Cables1(affects peak height velocity in puberty, essential for progression through mitosis)
-Caskin1
-PLEKHA1
-PXMP3(knockout causes skeletal defects)
-Cyp20a1
-L3MBTL3
-PENK
-RBBP8
-ADYC-POMC(variant associated with increased body mass index, reduced pubertal growth and earlier puberty)
-DEF6
-RPS20
-c17orf67(affects peak height velocity in puberty)
-SOCS2(negatively regulates HGH/IGF-1 pathway, estrogen induces SOCS2 expression,KO increases height, affects peak height velocity in puberty, decreased by JAK-STAT pathway in response to GH)
-ZNF678(may be involved in transcriptional regulation)
-IHH(essential for patterning events during development, mouse knockout causes skeletal defects)
-DYM(may have a role in protein digestion or proteoglycan metabolism, KO decreases height)
-VDR(Mediates the action of Vitamin D3, knockout decreases height)
-DBP (GC)(binds to the promoters of genes like CYP2A4)
-CYP2R1(involved with Vitamin D2 and D3)
-CYP24A1(degrades the active form of Vitamin D3)
-CASR(senses changes in calcium concentration)
-ALPL(Involved with Alkaline Phosphatase)
-PTH
-PTHIH
-GCKR(gain of function causes short stature)
-LCORL(affects birth weight as well, affects peak height during infancy)
-PTHLH(encodes PTHrP, KO reduces height)
-IGF1(overexpression can cause overgrowth postnatally)
-IGF1R(KO reduces height)
-IGF2R(KO increases height by increasing circulating IGF2 levels)
-LHX3-QSOX2(LHX3 is a transcription factor involved in prolactin promotion and plays a role in the pituitary gland, QSOX2 is involved in oxidation)
-PLIN1(involved in liposis of intracellular lipid deposits)
-PLIN4(involved in liposis of intracellular lipid deposits)
-PRKZC
-MFAP2
-HTR1D(receptor for seratonin)
-CLIC4(enhances TGF-Beta signaling, causes cell cycle arrest, inhibits c-Myc and p21, stabilizes pSmad2/3: TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3.)
-Smad4(Smad4 is required for the normal organization of the cartilage growth plate. states that knockout reduces height)
-NOG(KO increases height, supresses BMP signaling, overexpression blocks chondrogenesis, Conditional inactivation of noggin in the postnatal skeleton causes osteopenia., where Nog is knocked out in osteoblasts reduces height)
-NOS3
-GH1
-HNF4A
-IL6ST
-STAT2
-LATS2(Growth factor binding, TGF Beta protein complex)
-Cdh13
-BMP6(knockout causes growth defects)
-fbp2
-Frem1
-TGFBeta1(knockout reduces height)
-TGFBeta2(Mediates the effects of Ihh on hypertrophic chondrocyte differentiation and PTHrP expression, knockout reduces height, mutations associated with tall stature)
-PTCH1(mutations increase height, 9q22.3 microdeletion causes tall stature)
-CYP19A1(codes aromatase which converts testosterone to estrogen)
-EIF2AK3(Phosphorylates the alpha subunit of eukaryotic translation-initiation factor 2 (EIF2), leading to its inactivation and thus to a rapid reduction of translational initiation and repression of global protein synthesis. Can cause G1 growth arrest, knockout reduces height)
-FGFR4
-ID4(knockout reduces height)
-PML
-Fastkd2
-HMGA1(knockout causes growth defects)
-PPIF
-Ttll5
-PPARD(target gene of Vitamin D, knockout causes growth defects)
-RUNX2(induces IHH expression, knockout reduces height)
-RUNX3(increases bone growth when removed except when RUNX2 is also affected, overexpression increases chondrocyte hypertrophy and induces ectopic calcification)
-STC2(involved in calcium uptake, increase bone growth when removed, overexpression reduces height)
-Sulf1
-COL11A1(loss of function mutations cause short stature)
-GFPT2(biosynthesis of chondroitin sulfate)
-SERPINH1(matrix biosynthesis, mutations reduce height)
-GALNS(matrix degradation)
-ADAMS10(matrix degradation)
-TOP2A(DNA Synthesis)
-CENPW(mitotic spindle formation)
-CCNL1(affects birth length and birth weight)
-TACC3(mitotic spindle formation)
-E2F1(cell cycle progression, overexpression disrupts growth, if double KOed with E2F3 height growth is disturbed this is not present if only E2F1 is KOed)
-SLBP(translation of replication dependent histones)
-IGF2BP2(downregulated during growth plate senescence)
-IGF2BP3(upregulated during hypertrophic differentiation and downregulated during growth plate senescence, mutation associated with tall stature)
-IGF2BP1(removal of gene reduces bone length and drives growth plates to mineralization quicker)
-RSPO3(activates Wnt5a signaling)
-Wnt5a
-LTBP1(involved with TGFBeta and Growth factor binding)
-LTBP2(mutations associated with tall stature)
-Enpp2
-TRIM25
-Spin1
-LTA4H
-Ccdc1
-Ccdc88a
-LTBP3(KO decreases height, increases TGFBeta signaling)
-GLI2(mediates Ihh signaling in growth plate, knockout shortens long bones and delays ossification, growth factor binding, WNT protein binding)
-PDE11A
-TULP4
-GNA12
-Cdc42ep3
-DCC(receptor for netrin, binding with netrin-1 activates FAK signaling, regulates venous invasion)
-CDK6(knockout causes growth defects, essential for progression through mitosis)
-HOMER1
-Sssca1
-Dclk1
-Sox9
-Cdca7l
-PEX1
-Batf(controls differentiation of helper T-cells)
-BCL3(controls differentiation of helper T-cells)
-ANFKN1
-Syn3
-Atf7
-Akt1
-Ippk
-BCAS3
-MAF
-C19orf35
-NFATC4
-CRLF3
-ITM2A
-Esr1(estrogen receptor alpha, mutation resulted in increased height, knockout decreased height,disruption reduced growth hormone secretion)
-Esr2(knockout increases height in mice)
-Gas1(KO delays chondrogenesis and ossification, "Overexpression of Gas1 in limb micromass culture abolishes participation in cartilage formation"<-not sure what this means)
-MEF2C
-Nov(regulates actin cytoskeleton organization, antagonizes BMP-2)
-Nppc(2q37 translocation induces CNP overexpression and causes overgrowth)
-Npr3(limits CNP signaling in chondrocytes, knockout increases height)
-Npr2(codes the CNP receptor, knockout decreases height)
-Pcsk5(KO reduces height, upregulated by LSJL, PCSK5 encodes the proprotein convertase subtilisin/kexin type 5, the expression of which has been linked to developmental dynamics in mice, mutations associated with tall stature)
-Pds5B(KO reduces height)
-Pitx1(KO reduces height severely)
-SHOX2(knockout results in reduced height, overexpression increases height)
-VANGL2(mutations reduce body size)
-Twist1(knockout reduces height, overexpression increases height)
-TNS3(knockout increases resting zone but decreases chondrocyte proliferation with reduced height may promote stemness but reduce differentiation)
-TRIP11(mutations decrease height)
-Tacc3(Knockout reduces height)
-RPL5(Mutations cause short stature)
-TBX15(inhibits adipocyte differentiation, Knockout results in short stature)
-PRGK2(mediates the effects of cGMP, knockout causes dwarfism)
-FGF18(interacts with FGFR3, knockout increases height)
-NSD1(mutations can cause tall stature, knockout reduces height)
-T(knockout reduces height, may induce chondrogenic differentiation)
-BNC2(knockout reduces height)
-FANCC(mutations decrease height, knockout increases height)
-ZFP36L1(KO decreases height)
-NF1(KO decreases height)
-RPL13AP17
-ZFP638
-TBX2
-TBX4(involved in short mandible)
-ADCY5(affects birth length and weight)
-C18orf12
-Zfp76
-Zfp341
-Zfp462
-Zfp678
-Zfp510
-Zfat1
-OR2J3
-Fstl5
-Tns1
-ZFHX4
-NFIC
-PBX1
-PBS5B(
-Frs2(involved with PI3K, response to fibroblast growth factor)
-Ankrd60
-RNF135(mutations involving deletion can cause overgrowth)
-KCNJ2(KO decreases height)
-ADAMTS10(some mutations decrease height, other mutations increase height)
-Dlx5(OE reduces height)
-OASIS(Distinct mechanisms are responsible for osteopenia and growth retardation in OASIS-deficient mice., states that OASIS knockout reduces height, increases IGF-1 and GH levels, also called CREB3L1)
-TMED10
-Adamts3
-Atp5sl
-H19(H19 acts as a trans regulator of the imprinted gene network controlling growth in mice., conditional knockout of H19 increases height due to effect on IGF2 although the mutation needed to cause overgrowth is fairly specific)
-GPC3(Altered hematopoiesis in glypican-3-deficient mice results in decreased osteoclast differentiation and a delay in endochondral ossification., The loss of glypican-3 induces alterations in Wnt signaling., loss of function causes overgrowth, seems to slow down terminal differentiation, Mutations cause Simson-Golabi-Behmel syndrome, an X-linked disorder with pre- and post-natal overgrowth, [with] visceral and skeletal [defects]. GPC3 is a negative regulator of Hedgehog (HH), and hyperactivation of HH signaling leads to overgrowth, non-functional GPC3 may lead to increased IGF2 signalling)
-PHLDA2(Placental growth retardation due to loss of imprinting of Phlda2., total deletion results in overgrowth, partial deletion(loss of imprinting) reduces height, PHLDA2 overexpression can reduce height if expressed with Slc22a1l)
-KVDMR1(knockout reduces height)
-NEDD4L(partial deletion reduces height, regulator of sodium channels, could regulate TGF-Beta signaling, LSJL downregulates a few proteins in the NEDD4 family, NEDD4L binds to Smad7 which inhibits Smad's)
-Catsper4
-Dcaf12
-CERS6
-Gpc5
-GATAD1
-Slc22a5
-HPGD
-Gsdmc
-Slco1c1
-COIL
-EEF1A1P37
-ZHX2
-NAALADL2
-ATAD5
-OR2I1P
-PASK
-TGS1
-FBN2(mutation can cause overgrowth)
-GNAS1(loss of function mutation reduces height, plays a role in adenylate cyclase activation, gain of function results in abnormal fibrous overgrowth in the bone)
-TRA1(knockout reduces height,
-GPR30(knockout increases height)
-FUBP3
-Lyrm4
-LYAR
-TFAM
-TTF1
-GRB10(KO mice show embryo and placental overgrowth)
-FBLN5(ECM stabilizer, induces MMPs)
-MC4R(allele that is associated with greater height also associates with obesity, accelerates height during childhood)
-EGFR(KO reduces height)
-Girdin(Similar phenotypes of Girdin germ-line and conditional knockout mice indicate a crucial role for Girdin in the nestin lineage., Akt substrate, KO reduces height)
-PTEN(Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth., chondrogenic specific deletion causes overgrowth, haploinsufficiency leads to postnatal overgrowth but to reduced growth childhood leading to normal adult height)
-mitochondrian ATPase 6(Tall stature and progressive overweight in mitochondrial encephalopathy., mutation causes skeletal overgrowth)
-GHSR(GH secretagogue receptor gene polymorphisms are associated with stature throughout childhood., mutations associated with overgrowth but not necessarily adult tall stature, A genetic study of the ghrelin and growth hormone secretagogue receptor (GHSR) genes and stature., found no effect on adultheight)
-NSD1(mutations can cause tall stature, SOTOS syndrome)
-FMR1(mutations can cause tall stature, failure to express results in increased growth rate but reduced final stature)
There are several genes associated with methylation and histone related mechanisms. cGMP and GDF5's involvement in height is as expected. Also, of note no direct association to HGH(except for SOCS2) on height genes.
Each individual gene has only a small effect on height so maybe genes that have not been discovered yet influence proteins like BMP-2 and TGF-Beta.
Other studies used for genes:
Replication study of the association of SNPs in the LHX3-QSOX2 and IGF1 loci with adult height in the Japanese population; wide-ranging comparison of each SNP genotype distribution.
A Polymorphism in a gene encoding Perilipin 4 is associated with height but not with bone measures in individuals from the Framingham Osteoporosis Study.
KO(inhibit these):
FANCC
NOG
NPR3
RNF135
SOCS2
STC2
GPC3
IGF2R
GPR30
POMC
FGF21
FGFR3
FGFR1
ICR1
OE(stimulate these):
NPPC
PLAG1
SHOX2
Twist1
CNP
IGF2
IGF1
Akt1
CTGF(CCN2)
Mef2c[chondrocyte specific]
Biphasic Genes(genes need optimal quality):
HHIP
Zfp521
LCN2
Although, the genes involved in juvenile growth are not the same as the ones involved with reinvigorating growth. Analyzing the genes involved in juvenile growth can help illuminate the pathways involved in endochondral ossification like the HMGA2 gene which is associated with chromatin and let-7.
Identification of ten loci associated with height highlights new biological pathways in human growth.
"Ten newly identified and two previously reported loci were strongly associated with variation in height (P values from 4 × 10-7 to 8 × 10-22). Together, these 12 loci account for ~2% of the population variation in height. Individuals with ≤8 height-increasing alleles and ≥16 height-increasing alleles differ in height by ~3.5 cm. The newly identified loci, along with several additional loci with strongly suggestive associations, encompass both strong biological candidates and unexpected genes, and highlight several pathways (let-7 targets, chromatin remodeling proteins and Hedgehog signaling) as important regulators of human stature. These results expand the picture of the biological regulation of human height and of the genetic architecture of this classical complex trait."<-We've looked at IHH and chromatin remodeling. Let-7 is associated with osteoblast and type I collagen which means that Type I Collagen and osteoblasts may be more important than first thought.
The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature.
"Human height is considered highly heritable and correlated with certain disorders, such as type 2 diabetes and cancer. Despite environmental influences, genetic factors are known to play an important role in stature determination. A number of genetic determinants of adult height have already been established through genome wide association studies.
To examine 51 single nucleotide polymorphisms (SNPs) corresponding to the 46 previously reported genomic loci for height in 8,184 European American children with height measurements. We leveraged genotyping data from our ongoing GWA study of height variation in children in order to query the 51 SNPs in this pediatric cohort.
Sixteen of these SNPs yielded at least nominally significant association to height, representing fifteen different loci including EFEMP1-PNPT1, GPR126, C6orf173, SPAG17, Histone class 1, HLA class III and GDF5-UQCC. Other loci revealed no evidence for association, including HMGA1 and HMGA2. For the 16 associated variants, the genotype score explained 1.64% of the total variation for height z-score.
Among 46 loci that have been reported to associate with adult height to date, at least 15 also contribute to the determination of height in childhood."
Genome-wide association analysis identifies 20 loci that influence adult height.
"Adult height is a model polygenic trait, but there has been limited success in identifying the genes underlying its normal variation. To identify genetic variants influencing adult human height, we used genome-wide association data from 13,665 individuals and genotyped 39 variants in an additional 16,482 samples. We identified 20 variants associated with adult height (P < 5 × 10−7, with 10 reachingP < 1 × 10−10). Combined, the 20 SNPs explain ~3% of height variation, with a ~5 cm difference between the 6.2% of people with 17 or fewer ‘tall’ alleles compared to the 5.5% with 27 or more ‘tall’ alleles. The loci we identified implicate genes in Hedgehog signaling (IHH, HHIP, PTCH1), extracellular matrix (EFEMP1, ADAMTSL3, ACAN) and cancer (CDK6, HMGA2, DLEU7) pathways, and provide new insights into human growth and developmental processes. Finally, our results provide insights into the genetic architecture of a classic quantitative trait."
"Our findings clearly emphasize that Caucasians and East Asians do not share all of the genetic mechanisms for height"
List
-SNP in the 3′ UTR of the HMGA2 gene(Chromatin, Type I Collagen)(knockout causes growth defects)
-GDF5-UQCC(Chondrogenesis, may induce ectopic chondrogenesis thus could potentially be chondroinductive, KO reduces height, allele that increases height also reduces risk for osteoarthritis, affects peak height velocity during infancy)
-SH3GL3-ADAMTSL3( glycoprotein metalloprotease, regulates venous invasion)
-CDK6(associated with let-7, allele that associates with greater height also associates with greater risk of rheumatoid arthritis)
-CHCHD7-RDHE2
-ZBTB38(methyl-DNA-binding transcriptional repressor, methylation affects cellular senesence)
-GPR126(involved in myelination which is involved in nervous function thus nerves may be very important for height growth)
-HIST1H1D(compaction of chromatin into higher end structures, affects peak height velocity during infancy)
-HHIP(reduces Ihh signaling, HHIP overexpression reduces height, mouse knockout causes skeletal defects, affects peak height velocity during infancy)
-TRIP11-ATXN3(Thyroid Hormone Receptor, thyroid hormone is associated with IGF-1 receptors, mutations cause dwarfism)
-LIN28A
-Six6
-Sox5(involved in dwarfism)
-CDKAL1(affects birth length and weight)
-ADRB1(affects birth length and weight)
-LIN28B(associated with let-7)
-DOT1L(histone methyltransferase; histone modifies Sox9 transcription; associated with let-7, affects peak height velocity during puberty)
-PTCH1(SHH & IHH Receptor, knockout causes skeletal defects)
-ACAN(Aggrecan an ECM protein, KO reduces height)
-ADAMTS17(processes proteoglycans and cleaves ECM)
-ADAMTSL3(ECM-related, lacks metalloprotease and other distegrin-like domains unlike normal ADAMTS)
-SCMH1(maintains that transcriptively repressive state of some genes, modifies histone, mouse knockout causes skeletal defects)
-ANAPC13(controls progression through mitosis and G1 phase of the cell cycle)
-PRKG2(encodes the cGMP-dependent protein kinase II (cGKII); cGMP is associated with CNP and is involved in chondrocyte hypertrophy, upregulated in LSJL)
-NCAPG(required for conversion of interphase chromatin, essential for progression in mitosis)
-EFEMP1-PNPT1(EFEMP1 may play a role in cell adhesion and migration and may inhibit chondrocyte differentiation. EFEMP1 is also an ECM stabilizer. overexpression stimulates proliferation and supresses chondrocyte differentiation, knockout reduces body size)
-PLAG1(upregulates IGF-II, knockout causes growth defects, has a major effect on stature according to Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature.)
-SPAG17[upregulated by LSJL](maintains structural integrity of the sperm, may directly cause height or just be a correlation as it's important for parents to have SPAG17 but not yourself to maximize height)
-DLEU7(affects peak height velocity during infancy)
-Prkca
-SPAG1
-Slit3
-MOS
-Sox8(shortens rib length)
-Lyn
-CTNNB1
-AXIN2(Tgfb and Wnt involvement)
-Wnt4
-CHSY1(involved in TGFBeta signaling)
-FNDC3B(some diseases with decreased bone length have this involved)
-TRIOBP(involved with Beta-Catenin binding)
-BNC2(regulates cartilage differentiation, some mutations shorten ulna)
-WWP2(cartilage and chondrocyte development, affects PDGF signaling)
-Sp3(growth plate receptor signalling pathway)
-PLAGL1(mutations reduce height)
-Glt25d2
-C3orf47
-MTMR11
-C3orf63
-Cables1(affects peak height velocity in puberty, essential for progression through mitosis)
-Caskin1
-PLEKHA1
-PXMP3(knockout causes skeletal defects)
-Cyp20a1
-L3MBTL3
-PENK
-RBBP8
-ADYC-POMC(variant associated with increased body mass index, reduced pubertal growth and earlier puberty)
-DEF6
-RPS20
-c17orf67(affects peak height velocity in puberty)
-SOCS2(negatively regulates HGH/IGF-1 pathway, estrogen induces SOCS2 expression,KO increases height, affects peak height velocity in puberty, decreased by JAK-STAT pathway in response to GH)
-ZNF678(may be involved in transcriptional regulation)
-IHH(essential for patterning events during development, mouse knockout causes skeletal defects)
-DYM(may have a role in protein digestion or proteoglycan metabolism, KO decreases height)
-VDR(Mediates the action of Vitamin D3, knockout decreases height)
-DBP (GC)(binds to the promoters of genes like CYP2A4)
-CYP2R1(involved with Vitamin D2 and D3)
-CYP24A1(degrades the active form of Vitamin D3)
-CASR(senses changes in calcium concentration)
-ALPL(Involved with Alkaline Phosphatase)
-PTH
-PTHIH
-GCKR(gain of function causes short stature)
-LCORL(affects birth weight as well, affects peak height during infancy)
-PTHLH(encodes PTHrP, KO reduces height)
-IGF1(overexpression can cause overgrowth postnatally)
-IGF1R(KO reduces height)
-IGF2R(KO increases height by increasing circulating IGF2 levels)
-LHX3-QSOX2(LHX3 is a transcription factor involved in prolactin promotion and plays a role in the pituitary gland, QSOX2 is involved in oxidation)
-PLIN1(involved in liposis of intracellular lipid deposits)
-PLIN4(involved in liposis of intracellular lipid deposits)
-PRKZC
-MFAP2
-HTR1D(receptor for seratonin)
-CLIC4(enhances TGF-Beta signaling, causes cell cycle arrest, inhibits c-Myc and p21, stabilizes pSmad2/3: TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3.)
-Smad4(Smad4 is required for the normal organization of the cartilage growth plate. states that knockout reduces height)
-NOG(KO increases height, supresses BMP signaling, overexpression blocks chondrogenesis, Conditional inactivation of noggin in the postnatal skeleton causes osteopenia., where Nog is knocked out in osteoblasts reduces height)
-NOS3
-GH1
-HNF4A
-IL6ST
-STAT2
-LATS2(Growth factor binding, TGF Beta protein complex)
-Cdh13
-BMP6(knockout causes growth defects)
-fbp2
-Frem1
-TGFBeta1(knockout reduces height)
-TGFBeta2(Mediates the effects of Ihh on hypertrophic chondrocyte differentiation and PTHrP expression, knockout reduces height, mutations associated with tall stature)
-PTCH1(mutations increase height, 9q22.3 microdeletion causes tall stature)
-CYP19A1(codes aromatase which converts testosterone to estrogen)
-EIF2AK3(Phosphorylates the alpha subunit of eukaryotic translation-initiation factor 2 (EIF2), leading to its inactivation and thus to a rapid reduction of translational initiation and repression of global protein synthesis. Can cause G1 growth arrest, knockout reduces height)
-FGFR4
-ID4(knockout reduces height)
-PML
-Fastkd2
-HMGA1(knockout causes growth defects)
-PPIF
-Ttll5
-PPARD(target gene of Vitamin D, knockout causes growth defects)
-RUNX2(induces IHH expression, knockout reduces height)
-RUNX3(increases bone growth when removed except when RUNX2 is also affected, overexpression increases chondrocyte hypertrophy and induces ectopic calcification)
-STC2(involved in calcium uptake, increase bone growth when removed, overexpression reduces height)
-Sulf1
-COL11A1(loss of function mutations cause short stature)
-GFPT2(biosynthesis of chondroitin sulfate)
-SERPINH1(matrix biosynthesis, mutations reduce height)
-GALNS(matrix degradation)
-ADAMS10(matrix degradation)
-TOP2A(DNA Synthesis)
-CENPW(mitotic spindle formation)
-CCNL1(affects birth length and birth weight)
-TACC3(mitotic spindle formation)
-E2F1(cell cycle progression, overexpression disrupts growth, if double KOed with E2F3 height growth is disturbed this is not present if only E2F1 is KOed)
-SLBP(translation of replication dependent histones)
-IGF2BP2(downregulated during growth plate senescence)
-IGF2BP3(upregulated during hypertrophic differentiation and downregulated during growth plate senescence, mutation associated with tall stature)
-IGF2BP1(removal of gene reduces bone length and drives growth plates to mineralization quicker)
-RSPO3(activates Wnt5a signaling)
-Wnt5a
-LTBP1(involved with TGFBeta and Growth factor binding)
-LTBP2(mutations associated with tall stature)
-Enpp2
-TRIM25
-Spin1
-LTA4H
-Ccdc1
-Ccdc88a
-LTBP3(KO decreases height, increases TGFBeta signaling)
-GLI2(mediates Ihh signaling in growth plate, knockout shortens long bones and delays ossification, growth factor binding, WNT protein binding)
-PDE11A
-TULP4
-GNA12
-Cdc42ep3
-DCC(receptor for netrin, binding with netrin-1 activates FAK signaling, regulates venous invasion)
-CDK6(knockout causes growth defects, essential for progression through mitosis)
-HOMER1
-Sssca1
-Dclk1
-Sox9
-Cdca7l
-PEX1
-Batf(controls differentiation of helper T-cells)
-BCL3(controls differentiation of helper T-cells)
-ANFKN1
-Syn3
-Atf7
-Akt1
-Ippk
-BCAS3
-MAF
-C19orf35
-NFATC4
-CRLF3
-ITM2A
-Esr1(estrogen receptor alpha, mutation resulted in increased height, knockout decreased height,disruption reduced growth hormone secretion)
-Esr2(knockout increases height in mice)
-Gas1(KO delays chondrogenesis and ossification, "Overexpression of Gas1 in limb micromass culture abolishes participation in cartilage formation"<-not sure what this means)
-MEF2C
-Nov(regulates actin cytoskeleton organization, antagonizes BMP-2)
-Nppc(2q37 translocation induces CNP overexpression and causes overgrowth)
-Npr3(limits CNP signaling in chondrocytes, knockout increases height)
-Npr2(codes the CNP receptor, knockout decreases height)
-Pcsk5(KO reduces height, upregulated by LSJL, PCSK5 encodes the proprotein convertase subtilisin/kexin type 5, the expression of which has been linked to developmental dynamics in mice, mutations associated with tall stature)
-Pds5B(KO reduces height)
-Pitx1(KO reduces height severely)
-SHOX2(knockout results in reduced height, overexpression increases height)
-VANGL2(mutations reduce body size)
-Twist1(knockout reduces height, overexpression increases height)
-TNS3(knockout increases resting zone but decreases chondrocyte proliferation with reduced height may promote stemness but reduce differentiation)
-TRIP11(mutations decrease height)
-Tacc3(Knockout reduces height)
-RPL5(Mutations cause short stature)
-TBX15(inhibits adipocyte differentiation, Knockout results in short stature)
-PRGK2(mediates the effects of cGMP, knockout causes dwarfism)
-FGF18(interacts with FGFR3, knockout increases height)
-NSD1(mutations can cause tall stature, knockout reduces height)
-T(knockout reduces height, may induce chondrogenic differentiation)
-BNC2(knockout reduces height)
-FANCC(mutations decrease height, knockout increases height)
-ZFP36L1(KO decreases height)
-NF1(KO decreases height)
-RPL13AP17
-ZFP638
-TBX2
-TBX4(involved in short mandible)
-ADCY5(affects birth length and weight)
-C18orf12
-Zfp76
-Zfp341
-Zfp462
-Zfp678
-Zfp510
-Zfat1
-OR2J3
-Fstl5
-Tns1
-ZFHX4
-NFIC
-PBX1
-PBS5B(
-Frs2(involved with PI3K, response to fibroblast growth factor)
-Ankrd60
-RNF135(mutations involving deletion can cause overgrowth)
-KCNJ2(KO decreases height)
-ADAMTS10(some mutations decrease height, other mutations increase height)
-Dlx5(OE reduces height)
-OASIS(Distinct mechanisms are responsible for osteopenia and growth retardation in OASIS-deficient mice., states that OASIS knockout reduces height, increases IGF-1 and GH levels, also called CREB3L1)
-TMED10
-Adamts3
-Atp5sl
-WNT6(hh signaling)
-WNT9a(hh signaling)
-WNT10A(hh signaling)
-WNT3A(hh signaling)
-FBXW11(hh signaling)
-MAP2K3
-CREBBP(autosomal dominant CREBBP and EP300 genes cause short stature)
-EP300(autosomal dominant CREBBP and EP300 genes cause short stature)
-Acbd4
-MAPK3(variant associated with additional pubertal growth and earlier monarch, aka ERK1, adolescent height-increasing allele (G) at rs4788196 on 16p11.2 correlated with decreased expression of MAPK3, consistent with previous studies linking deactivation of the gene with increased bone growth in mice)
-DHRS1(Gamma-hexachlorocyclohexane degradation)
-ATP13A2(Folate biosynthesis)
-PCK2(Citrate Cycle)
-TAP1(antigen sensing)
-TAP2(antigen sensing)
-DGKE
-Dgkh
-CS(Citrate Synthase)
-RTF1
-RYBP
-PAX3(loss of function associated with short stature)
-Antxr1
-Arse
-SBNO1
-Il1r1(knockout increases height in mice)
-Lepr(knockin increases height in mice, Leptin receptor, mutations result in delayed puberty and reduced final height)
-Lep(Leptin, mutations result in delayed puberty and reduced final height)
-Pomc(knockout increases height in mice, gain of function causes short stature)
-SF3B4(also known as SV2A, contributes to the regognition of nRNA intron's branch points. Binds the BMPR-IA serine/threonine kinase receptor, affects peak height velocity during infancy and puberty)
-F3B4(binds BMPR-IA and inhibits BMP-induced SMAD1/5/8 pathways)
-Kif3a(Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis, states that Kif3a knockout reduces growth plate organization and may reduce height; Mechanosensing by the primary cilium: deletion of Kif3A reduces bone formation due to loading. states Kif3a may be involved in mechanotransduction, The study identified Pkd1 as another gene involved in mechanotransduction)
-STK36(relates to Ihh signaling, Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus., upregulated in LSJL, knockout reduces height)-WNT9a(hh signaling)
-WNT10A(hh signaling)
-WNT3A(hh signaling)
-FBXW11(hh signaling)
-MAP2K3
-CREBBP(autosomal dominant CREBBP and EP300 genes cause short stature)
-EP300(autosomal dominant CREBBP and EP300 genes cause short stature)
-Acbd4
-MAPK3(variant associated with additional pubertal growth and earlier monarch, aka ERK1, adolescent height-increasing allele (G) at rs4788196 on 16p11.2 correlated with decreased expression of MAPK3, consistent with previous studies linking deactivation of the gene with increased bone growth in mice)
-DHRS1(Gamma-hexachlorocyclohexane degradation)
-ATP13A2(Folate biosynthesis)
-PCK2(Citrate Cycle)
-TAP1(antigen sensing)
-TAP2(antigen sensing)
-DGKE
-Dgkh
-CS(Citrate Synthase)
-RTF1
-RYBP
-PAX3(loss of function associated with short stature)
-Antxr1
-Arse
-SBNO1
-Il1r1(knockout increases height in mice)
-Lepr(knockin increases height in mice, Leptin receptor, mutations result in delayed puberty and reduced final height)
-Lep(Leptin, mutations result in delayed puberty and reduced final height)
-Pomc(knockout increases height in mice, gain of function causes short stature)
-SF3B4(also known as SV2A, contributes to the regognition of nRNA intron's branch points. Binds the BMPR-IA serine/threonine kinase receptor, affects peak height velocity during infancy and puberty)
-F3B4(binds BMPR-IA and inhibits BMP-induced SMAD1/5/8 pathways)
-Kif3a(Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis, states that Kif3a knockout reduces growth plate organization and may reduce height; Mechanosensing by the primary cilium: deletion of Kif3A reduces bone formation due to loading. states Kif3a may be involved in mechanotransduction, The study identified Pkd1 as another gene involved in mechanotransduction)
-H19(H19 acts as a trans regulator of the imprinted gene network controlling growth in mice., conditional knockout of H19 increases height due to effect on IGF2 although the mutation needed to cause overgrowth is fairly specific)
-GPC3(Altered hematopoiesis in glypican-3-deficient mice results in decreased osteoclast differentiation and a delay in endochondral ossification., The loss of glypican-3 induces alterations in Wnt signaling., loss of function causes overgrowth, seems to slow down terminal differentiation, Mutations cause Simson-Golabi-Behmel syndrome, an X-linked disorder with pre- and post-natal overgrowth, [with] visceral and skeletal [defects]. GPC3 is a negative regulator of Hedgehog (HH), and hyperactivation of HH signaling leads to overgrowth, non-functional GPC3 may lead to increased IGF2 signalling)
-PHLDA2(Placental growth retardation due to loss of imprinting of Phlda2., total deletion results in overgrowth, partial deletion(loss of imprinting) reduces height, PHLDA2 overexpression can reduce height if expressed with Slc22a1l)
-KVDMR1(knockout reduces height)
-NEDD4L(partial deletion reduces height, regulator of sodium channels, could regulate TGF-Beta signaling, LSJL downregulates a few proteins in the NEDD4 family, NEDD4L binds to Smad7 which inhibits Smad's)
-Catsper4
-Dcaf12
-CERS6
-Gpc5
-GATAD1
-Slc22a5
-HPGD
-Gsdmc
-Slco1c1
-COIL
-EEF1A1P37
-ZHX2
-NAALADL2
-ATAD5
-OR2I1P
-PASK
-TGS1
-FBN2(mutation can cause overgrowth)
-GNAS1(loss of function mutation reduces height, plays a role in adenylate cyclase activation, gain of function results in abnormal fibrous overgrowth in the bone)
-TRA1(knockout reduces height,
-GPR30(knockout increases height)
-FUBP3
-Lyrm4
-LYAR
-TFAM
-TTF1
-GRB10(KO mice show embryo and placental overgrowth)
-FBLN5(ECM stabilizer, induces MMPs)
-MC4R(allele that is associated with greater height also associates with obesity, accelerates height during childhood)
-EGFR(KO reduces height)
-Girdin(Similar phenotypes of Girdin germ-line and conditional knockout mice indicate a crucial role for Girdin in the nestin lineage., Akt substrate, KO reduces height)
-PTEN(Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth., chondrogenic specific deletion causes overgrowth, haploinsufficiency leads to postnatal overgrowth but to reduced growth childhood leading to normal adult height)
-mitochondrian ATPase 6(Tall stature and progressive overweight in mitochondrial encephalopathy., mutation causes skeletal overgrowth)
-GHSR(GH secretagogue receptor gene polymorphisms are associated with stature throughout childhood., mutations associated with overgrowth but not necessarily adult tall stature, A genetic study of the ghrelin and growth hormone secretagogue receptor (GHSR) genes and stature., found no effect on adultheight)
-NSD1(mutations can cause tall stature, SOTOS syndrome)
-FMR1(mutations can cause tall stature, failure to express results in increased growth rate but reduced final stature)
-HDLBP(downregulated by LSJL)
-SMPD2(downregulated by LSJL)
-GPR133
-BMP2
-FEN1
-col27a1
-c6orf106
-DNM3
-TMEM132C
-ETV6
-JAZF1
-LOC387103
-Nkx2.1
-Pappa
-Bod1
-Pdgfra
-Polg
-Rab40c
-Wdr60
-Ext1
-Hla-B
-Supt3h
-Dis3l2
-Dnajc27
-MAML2(transcriptional co-activator of Notch proteins)
-MYO16
-C16orf55
-SMPD2(downregulated by LSJL)
-GPR133
-BMP2
-FEN1
-col27a1
-c6orf106
-DNM3
-TMEM132C
-ETV6
-JAZF1
-LOC387103
-Nkx2.1
-Pappa
-Bod1
-Pdgfra
-Polg
-Rab40c
-Wdr60
-Ext1
-Hla-B
-Supt3h
-Dis3l2
-Dnajc27
-MAML2(transcriptional co-activator of Notch proteins)
-MYO16
-C16orf55
There are several genes associated with methylation and histone related mechanisms. cGMP and GDF5's involvement in height is as expected. Also, of note no direct association to HGH(except for SOCS2) on height genes.
Each individual gene has only a small effect on height so maybe genes that have not been discovered yet influence proteins like BMP-2 and TGF-Beta.
Other studies used for genes:
Replication study of the association of SNPs in the LHX3-QSOX2 and IGF1 loci with adult height in the Japanese population; wide-ranging comparison of each SNP genotype distribution.
A Polymorphism in a gene encoding Perilipin 4 is associated with height but not with bone measures in individuals from the Framingham Osteoporosis Study.
<-Important study
Meta-Analysis of Genome-Wide Scans for Human Adult Stature Identifies Novel Loci and Associations with Measures of Skeletal Frame Size
Genome-Wide Association Study Identified CNP12587 Region Underlying Height Variation in Chinese Females.
Overgrowth.
Gene-based copy number variation study reveals a microdeletion at 12q24 that influences height in the Korean population.
Functions and physiological roles of two types of estrogen receptors, ERα and ERβ, identified by estrogen receptor knockout mouse.
Genetic analysis of tall stature.
Genetic regulation of the growth plate.
PIK3CA, a hotspot for postzygotic mutations in nonhereditary overgrowth syndromes
"finding of 12.5% of the variance in heritability of body height (by 18GWAS loci)"
Using height association studies to gain insights into human idiopathic short and syndromic stature phenotypes.
Synthesizing genome-wide association studies and expression microarray reveals novel genes that act in the human growth plate to modulate height.
"Many of the 78 implicated genes participate in molecular pathways that are known to be important for growth plate chondrogenesis in the mouse. A large number participate in signaling pathways that regulate growth plate chondrocyte proliferation and differentiation, including Indian hedgehog, parathyroid hormone-related peptide, bone morphogenetic protein / transforming growth factor-beta superfamily, C-type natriuretic peptide, fibroblast growth factor, and insulin-like growth factor signaling"
New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism.
Reaching new heights: insights into the genetics of human stature
Human height genes and cancer.
"TP53, ESR1, HNF4A and MYC are main network hubs [for height increase]."
Genome-wide association study in Han Chinese identifies three novel loci for human height.
Rare copy number variants are a common cause of short stature.
Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity.
Genetics of obesity and overgrowth syndromes
Genetic Determinants of Height Growth Assessed Longitudinally from Infancy to Adulthood in the Northern Finland Birth Cohort 1966
Human growth is associated with distinct patterns of gene expression in evolutionarily conserved networks.
Common DNA variants predict tall stature in Europeans.
Defining the role of common variation in the genomic and biological architecture of adult human height.
Data from jax.org was used as well.
Meta-Analysis of Genome-Wide Scans for Human Adult Stature Identifies Novel Loci and Associations with Measures of Skeletal Frame Size
Genome-Wide Association Study Identified CNP12587 Region Underlying Height Variation in Chinese Females.
Overgrowth.
Gene-based copy number variation study reveals a microdeletion at 12q24 that influences height in the Korean population.
Genetic analysis of tall stature.
Genetic regulation of the growth plate.
Genome-Wide Association Studies of Skeletal Phenotypes: What We Have Learned and Where We Are Headed.
Using height association studies to gain insights into human idiopathic short and syndromic stature phenotypes.
"at the extreme tall end of the height distribution, phenotypes were consistent with the segregation of common DNA polymorphisms, each with a weak effect on height. "
"Many of the 78 implicated genes participate in molecular pathways that are known to be important for growth plate chondrogenesis in the mouse. A large number participate in signaling pathways that regulate growth plate chondrocyte proliferation and differentiation, including Indian hedgehog, parathyroid hormone-related peptide, bone morphogenetic protein / transforming growth factor-beta superfamily, C-type natriuretic peptide, fibroblast growth factor, and insulin-like growth factor signaling"
New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism.
Finding missing heritability in less significant Loci and allelic heterogeneity: genetic variation in human height.
"including less significant loci (i.e., p-value<5×10(-4)) and accounting for effects of allelic heterogeneity substantially improved the variance explained in height."
"Height was normally distributed in both males (N = 565) and females (N = 739) with larger variability observed in males"<-Sox9 is a sex linked trait.
Reaching new heights: insights into the genetics of human stature
Human height genes and cancer.
"TP53, ESR1, HNF4A and MYC are main network hubs [for height increase]."
Genome-wide association study in Han Chinese identifies three novel loci for human height.
Rare copy number variants are a common cause of short stature.
Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity.
Genetics of obesity and overgrowth syndromes
Genetic Determinants of Height Growth Assessed Longitudinally from Infancy to Adulthood in the Northern Finland Birth Cohort 1966
Human growth is associated with distinct patterns of gene expression in evolutionarily conserved networks.
Common DNA variants predict tall stature in Europeans.
Defining the role of common variation in the genomic and biological architecture of adult human height.
Labels:
height genes
Monday, August 27, 2012
Grow Taller with Horny Goat Weed?
Testosterone is an inhibitor of myostatin and myostatin inhibits cellular proliferation possibly through DNA methylation. Myostatin has been linked to height growth. Horny Goat weed is a substance that is readily available on sites like Amazon: HORNY GOAT WEED - 90 Capsules Male Performance Enhancement Aphrodis...ENERZIN (Make sure Icariin is in the ingredients). BMP-2 is also very important for initial chondrogenesis and Icariin increases BMP-2 mRNA expression so it can help you grow taller. Note that BMP-2 accelerates terminal differentiation in existing growth plates so you don't want to take Icariin if you are not skeletally mature. Starting chondrogenesis requires both TGF-Beta and BMP-2 and the LSJL gene expression studies don't clearly show an increase in BMP-2. So if LSJL does not increase BMP-2 levels then Icariin may be what's needed to help initiate LSJL results(but only for individuals with mature skeletons).
(Make sure Icariin is in the ingredients). BMP-2 is also very important for initial chondrogenesis and Icariin increases BMP-2 mRNA expression so it can help you grow taller. Note that BMP-2 accelerates terminal differentiation in existing growth plates so you don't want to take Icariin if you are not skeletally mature. Starting chondrogenesis requires both TGF-Beta and BMP-2 and the LSJL gene expression studies don't clearly show an increase in BMP-2. So if LSJL does not increase BMP-2 levels then Icariin may be what's needed to help initiate LSJL results(but only for individuals with mature skeletons).
Turns out Icariin may be pro-chondrogenic after all:
Icariin: a potential promoting compound for cartilage tissue engineering.
"Icariin was added into cell-hydrogel constructs derived from neonatal rabbit chondrocytes and collagen type I. The effects of icariin-added cell-hydrogel constructs on the restoration of supercritical-sized osteochondral defects of adult rabbit were investigated by histological observation. The cell-hydrogel constructs without Icariin were set for controls.
Icariin obviously up-regulate the expressions included aggrecan, sox9, and collagen type II of seeded chondrocytes form 99.7% to 248%[That's huge]. It increases the synthesis of glycosaminoglycan and collagen type II about fourfolds to fivefolds from week 1 to week 4, and accelerates the formation of chondroid tissue in the cell-hydrogel constructs. [Icariin] improves the restoration efficiency of super critical-sized osteochondral defects in adult rabbit model, and enhances the integration of new-formed cartilage with subchondral bone.
Icariin can be a substitute for the use of some growth factors."
"Herb Epimedium (HEP) [contains Icariin]"
"Icariin could regulate the anabolism of osteoblasts through the up-regulation of BMP-4, BMP-2 and SMAD4 expression"
"the osteoinductive effect of icariin in vivo might be expressed through the process of endochondral ossification"
"icariin [may] be a promoter of cartilage tissue engineering"
"the addition of icariin could help to increase the synthesis of cartilage-specific matrixes of the seeded chondrocytes, promote the formation of chondroid tissue"
It's possible that Icariin promotes the induction of Sox9 itself which would make it a very promising chondroinductive supplement but even if Icariin only enhances Sox9, COL2A1, and Aggrecan it may still help increase height in existing growth plates or help induce new growth plates with LSJL.
[Effect of icariin on the mRNA expressions of Cbfalpha1, BMP2, BMP4 in rat osteoblasts].
"Primary rat osteoblastic cells were obtained. The passage 3-5 cells were treated with icariin at the concentration of 0 mol/L, 10(-8) mol/L, 10(-7) mol/L, 10(-6) mol/L, 10(-5) mol/L, 10(-4) mol/L for 24 h, 48 h, 72 h, and the proliferation of the cells was measured.
Icariin showed no effect on the proliferation of osteoblasts, but improved ALP activity. The Cbfalpha1, BMP2, BMP4 mRNA were significantly upregulated after icariin treatment."
Icariin can increase BMP-2 expression in osteoblasts that BMP-2 expression may help induce chondrogenic differentiation in mesenchymal stem cells.
Analysis of the effects of androgens and training on myostatin propeptide and follistatin concentrations in blood and skeletal muscle using highly sensitive Immuno PCR.
"Myostatin propeptide (MYOPRO) and follistatin (FOLLI) are potent myostatin inhibitors. we analysed effects of training and androgens on MYOPRO and FOLLI concentrations in blood and skeletal muscle using Immuno PCR. Young healthy males performed either a 3-month endurance training or a strength training. Training did not significantly affect MYOPRO and FOLLI concentrations in serum and muscle. Blood samples of tetraplegic patients, untrained volunteers and bodybuilders were analysed. MYOPRO was significantly increased exclusively in the bodybuilder group[but it could be that the high levels of MYOPRO are causing the increased muscle mass rather than the other way around, longitudinal studies would have to performed]. In orchiectomised rats MYOPRO increased in blood and muscle after treatment with testosterone. Moderate training does not affect the concentrations of MYOPRO to FOLLI. In contrast androgen treatment results in a significant increase of MYOPRO in skeletal muscle and serum."
Testosterone inhibits myostatin through MYOPRO and horny goat weed(icariin) mimcs the effects of testosterone.
"there is only a moderate inter-individual variability with respect to the measured MYOPRO and FOLLI serum levels. In our study the serum concentrations varied between 6 and 10 ng/ml for FOLLI and between 35 and 50 pg/ml for MYOPRO."<-providing evidence that it's muscle mass that reduces concetration of myostatin inhibitors.
"no major effects of long term training on myostatin or its propeptide could be detected."<-so it's the muscle itself that inhibits myostatin and not the training.
Also note that there was no difference between the extremely low muscle mass group and the control group so there could be other factors. The scientists mention steroids.
The testosterone mimetic properties of icariin
"Forty-eight healthy male Sprague-Dawley rats at the age of 15 months were randomly divided into four groups with 12 rats each: the control group (C), the model group (M), the icariin group (ICA) and the testosterone group (T). The reproductive system was damaged by cyclophosphamide (intraperitoneal injection, 20 mg/kg day) for 5 consecutive days for groups M, ICA and T, at the sixth day, ICA (gastric gavage, 200 mg/kg
day) for 5 consecutive days for groups M, ICA and T, at the sixth day, ICA (gastric gavage, 200 mg/kg day) for the ICA group and sterandryl (subcutaneous injection, 5 mg/rat
day) for the ICA group and sterandryl (subcutaneous injection, 5 mg/rat day) for the T group for 7 consecutive days, respectively. The levels of serum testosterone, luteinizing hormone (LH), follicle stimulating hormone (FSH), serum bone Gla-protein (BGP) and tartrate-resistant acid phosphatase activity in serum (StrACP) were determined.
day) for the T group for 7 consecutive days, respectively. The levels of serum testosterone, luteinizing hormone (LH), follicle stimulating hormone (FSH), serum bone Gla-protein (BGP) and tartrate-resistant acid phosphatase activity in serum (StrACP) were determined.
(1) Icariin improved increased the circulating levels of testosterone.
(2) Icariin treatment also improved the steady-state serum BGP and might have promoted bone formation. At the same time, it decreased the serum levels of StrACP and might have reduced the bone resorption.
Icariin has testosterone mimetic properties."
Icariin "stimulates the differentiation of osteoblasts and increases the activity of alkaline phosphatase (ALP) and the expression of I-collagen protein in rats and promotes bone formation"<-this does not mean that Icariin is anti-chondrogenic.
The increase in Testosterone by Icariin was insane, 3 fold versus the control group and only 1/3rd of the direct testosterone group.
Icariin promotes bone formation. BGP is also known as osteocalcin which is secreted by osteoblasts so it's used as a measure of bone formation. Therefore, Icariin increase bone formation. It's Icariin's usage as a testosterone mimetic that is intriguing however as myostatin inhibition has the most potential usage during endochondral ossification.
[Experimental study of icariin in inducing bone marrow mesenchymal stem cell differentiation]
"To research the effect of icariin (ICA) on the proliferation and differentiation of bone mesenchymal stem cells (BMSCs) and to study its influence on the expressions of transforming growth factor-beta1 (TGF-beta1) and bone morphogenetic protein-2 (BMP-2) in the progress of BMSCs differentiating into osteoblast, for providing an experimental evidence to explain the mechanism of ICA.
After the most effective concentration of ICA for promoting the differentiation of BMSCs into osteoblast was judged with the indices like alkaline phosphatase (ALP), etc., a grouped experiment was conducted for the sake of studying the effect and mechanism of ICA in its process of inducing BMSCs differentiation into osteoblast through detecting expression of ALP and calcium nodes, as well as the expressions of TGF-beta1, and BMP-2 with ELISA.
The most effective concentration of the ICA on the BMSCs differentiation was judged as 1 x 10(-9) mol/L, ICA of that concentration showed effects in enhancing the expressions of osteoblast-indices and increasing the secretion of TGF-beta1, and BMP-2. the increase of TGF-beta1, and BMP-2 was revealed in all the groups, in which ICA showed its influence visibly.
ICA could promote the differentiation of BMSCs into osteoblast; the up-regulation of TGF-beta1, and BMP-2 expressions is possibly one of the action mechanisms of various interventional drugs in their differentiation promoting progress."<-Note this shows that it increased BMP-2 secretion so definitely potential to induce chondrogenic differentiation. The study mentions encouraging the differentiation of MSCs into osteoblasts but both TGF-Beta1 and BMP-2 are pro-chondrogenic.
On the bone resorption inhibition(and remember bone resorption may be an important part of endochondral ossification):
Icariin inhibits osteoclast differentiation and bone resorption by suppression of MAPKs/NF-kappaB regulated HIF-1alpha and PGE(2) synthesis.
"We examined the detail molecular mechanisms of icariin on lipopolysaccharide (LPS)-induced osteolysis. Icariin [inhibits] osteoclast differentiation and bone resorption by suppressing MAPKs/NF-kappaB regulated HIF-1alpha and PGE(2) synthesis[HIF-1alpha and the NF-kappaB pathway are both good for chondrogenesis/height growth]. After treatment with icariin, the activity of osteoclasts differentiation maker, tatrate resistances acid phosphatease (TRAP), significantly decreased at the concentration of 10(-8)M. Icariin (10(-8)M) reduced the size of LPS-induced osteoclasts formation, and diminished their TRAP and acid phosphatease (ACP) activity without inhibition of cell viability. Icariin also inhibited LPS-induced bone resorption and interleukin-6 (IL-6)[at 10(-8)M], and tumor necrosis factor-alpha (TNF-alpha) expression. The gene expression of osteoprotegerin (OPG) was up-regulated, while receptor activator of NF-kappaB ligand (RANKL) was down-regulated. Icariin also inhibited the synthesis of cyclo-oxygenase type-2 (COX-2) and prostaglandin E(2) (PGE(2)). In addition, icariin had a dominant repression effect on LPS-induced hypoxia inducible factor-1alpha (HIF-1alpha) expression of osteoclasts. On osteoclasts, icariin suppresses LPS-mediated activation of the p38 and JNK; while on the osteoblasts, icariin reduced the LPS-induced activation of ERK1/2 and I-kappa-B-alpha (IkappaBalpha), but increased the activation of p38. Icariin has an in vitro inhibitory effects on osteoclasts differentiation that can prevent inflammatory bone loss. Icariin inhibited LPS-induced osteoclastogenesis program by suppressing activation of the p38 and JNK pathway."
"In macrophages, LPS induces hypoxia-inducible factor-1α (HIF-1α) protein accumulation under normoxic condition"<-this could be important for us as hypoxia is pro-chondrogenic.
Icariin has inhibitory effects on hypoxia in this study and chondrocytes like to grow in low oxygen environments but the inhibitory effects may be specific to LPS.
According to [Comparative study on effect of icariin and genistein on proliferation and mineralization of osteoblasts in vitro]., Icariin may also increase bFGF and IGF-1.
Icariin Promotes Extracellular Matrix Synthesis and Gene Expression of Chondrocytes In Vitro.
"Rabbit chondrocytes were isolated from articular cartilage and cultured in vitro with different concentrations of icariin. Icariin at concentrations under 1 × 10(-5) m showed low cytotoxicity toward chondrocytes, but icariin at 5 × 10(-5) m inhibited the proliferation of chondrocytes. Icariin hardly affected the cell morphology with concentrations ranging from 1 × 10(-7) m to 5 × 10(-5) m. the higher concentration of icariin produced more extracellular matrix (ECM) synthesis and expression of chondrogenesis genes of chondrocytes. Indeed, the promotion of icariin on the synthesis of glycosaminoglycans (GAGs) and collagen of chondrocytes, and finally exerting a potent chondrogenic effect, might be due to its ability to up-regulate the expression of aggrecan, collagen II and Sox9 genes and to down-regulate the expression of the collagen I gene of chondrocytes. Icariin [is] an effective accelerant for chondrogenesis. 1 × 10(-5) m may be a suitable concentration of icariin with chondrogenic effect for tissue engineering"
"icariin media at a concentration of 5 × 10−5 m inhibited the proliferation of chondrocytes, it violently promoted GAG synthesis of cells."<-Whether this is beneficial to height growth is the question. If chondrocytes are not proliferating they are not approaching senescence.
So, looks like Horny Goat Weed may have all the makings of a supplement to help you grow taller except that it downregulates NF-KappaB, PGE2, IL-6, and HIF-alpha. It does increase p38.
Icariin: Does It Have An Osteoinductive Potential for Bone Tissue Engineering?
"Icariin, a typical flavonol glycoside, is considered to be the main active ingredient of the Herba Epimedii from which icariin has been successfully extracted. Icariin can be delivered locally by biomaterials and that it has an osteoinductive potential for bone tissue engineering. The osteoinductive potential of icariin could be attributed to its multiple functions in the musculoskeletal system which is involved in the regulation of multiple signaling pathways in anti-osteoporosis, osteogenesis, anti-osteoclastogenesis, chondrogenesis, angiogenesis, and anti-inflammation."
This paper mentions a study where cell-hydrogel construct delivery of icariin in rabbits increased cartilage formation.
"Icariin inhibited LPS-induced osteoclastogenesis by suppressing the activation of the p38 and JNK pathway"
"The effect of icariin on the synthesis of glycosaminoglycans (GAGs) and collagen of chondrocytes, and its potent chondrogenic effect, might be due to its ability to up-regulate the expression of aggrecan, collagen II, and Sox9 genes and to down-regulate the expression of the collagen I gene of chondrocytes"
" Icariin can up-regulate the expression of Sox9 and Cbfa1 in controlling osteogenesis and chondrogenesis."
Turns out Icariin may be pro-chondrogenic after all:
Icariin: a potential promoting compound for cartilage tissue engineering.
"Icariin was added into cell-hydrogel constructs derived from neonatal rabbit chondrocytes and collagen type I. The effects of icariin-added cell-hydrogel constructs on the restoration of supercritical-sized osteochondral defects of adult rabbit were investigated by histological observation. The cell-hydrogel constructs without Icariin were set for controls.
Icariin obviously up-regulate the expressions included aggrecan, sox9, and collagen type II of seeded chondrocytes form 99.7% to 248%[That's huge]. It increases the synthesis of glycosaminoglycan and collagen type II about fourfolds to fivefolds from week 1 to week 4, and accelerates the formation of chondroid tissue in the cell-hydrogel constructs. [Icariin] improves the restoration efficiency of super critical-sized osteochondral defects in adult rabbit model, and enhances the integration of new-formed cartilage with subchondral bone.
Icariin can be a substitute for the use of some growth factors."
"Herb Epimedium (HEP) [contains Icariin]"
"Icariin could regulate the anabolism of osteoblasts through the up-regulation of BMP-4, BMP-2 and SMAD4 expression"
"the osteoinductive effect of icariin in vivo might be expressed through the process of endochondral ossification"
"icariin [may] be a promoter of cartilage tissue engineering"
"the addition of icariin could help to increase the synthesis of cartilage-specific matrixes of the seeded chondrocytes, promote the formation of chondroid tissue"
It's possible that Icariin promotes the induction of Sox9 itself which would make it a very promising chondroinductive supplement but even if Icariin only enhances Sox9, COL2A1, and Aggrecan it may still help increase height in existing growth plates or help induce new growth plates with LSJL.
[Effect of icariin on the mRNA expressions of Cbfalpha1, BMP2, BMP4 in rat osteoblasts].
"Primary rat osteoblastic cells were obtained. The passage 3-5 cells were treated with icariin at the concentration of 0 mol/L, 10(-8) mol/L, 10(-7) mol/L, 10(-6) mol/L, 10(-5) mol/L, 10(-4) mol/L for 24 h, 48 h, 72 h, and the proliferation of the cells was measured.
Icariin showed no effect on the proliferation of osteoblasts, but improved ALP activity. The Cbfalpha1, BMP2, BMP4 mRNA were significantly upregulated after icariin treatment."
Icariin can increase BMP-2 expression in osteoblasts that BMP-2 expression may help induce chondrogenic differentiation in mesenchymal stem cells.
Analysis of the effects of androgens and training on myostatin propeptide and follistatin concentrations in blood and skeletal muscle using highly sensitive Immuno PCR.
"Myostatin propeptide (MYOPRO) and follistatin (FOLLI) are potent myostatin inhibitors. we analysed effects of training and androgens on MYOPRO and FOLLI concentrations in blood and skeletal muscle using Immuno PCR. Young healthy males performed either a 3-month endurance training or a strength training. Training did not significantly affect MYOPRO and FOLLI concentrations in serum and muscle. Blood samples of tetraplegic patients, untrained volunteers and bodybuilders were analysed. MYOPRO was significantly increased exclusively in the bodybuilder group[but it could be that the high levels of MYOPRO are causing the increased muscle mass rather than the other way around, longitudinal studies would have to performed]. In orchiectomised rats MYOPRO increased in blood and muscle after treatment with testosterone. Moderate training does not affect the concentrations of MYOPRO to FOLLI. In contrast androgen treatment results in a significant increase of MYOPRO in skeletal muscle and serum."
Testosterone inhibits myostatin through MYOPRO and horny goat weed(icariin) mimcs the effects of testosterone.
"there is only a moderate inter-individual variability with respect to the measured MYOPRO and FOLLI serum levels. In our study the serum concentrations varied between 6 and 10 ng/ml for FOLLI and between 35 and 50 pg/ml for MYOPRO."<-providing evidence that it's muscle mass that reduces concetration of myostatin inhibitors.
"no major effects of long term training on myostatin or its propeptide could be detected."<-so it's the muscle itself that inhibits myostatin and not the training.
Also note that there was no difference between the extremely low muscle mass group and the control group so there could be other factors. The scientists mention steroids.
The testosterone mimetic properties of icariin
"Forty-eight healthy male Sprague-Dawley rats at the age of 15 months were randomly divided into four groups with 12 rats each: the control group (C), the model group (M), the icariin group (ICA) and the testosterone group (T). The reproductive system was damaged by cyclophosphamide (intraperitoneal injection, 20 mg/kg
 day) for 5 consecutive days for groups M, ICA and T, at the sixth day, ICA (gastric gavage, 200 mg/kg
day) for 5 consecutive days for groups M, ICA and T, at the sixth day, ICA (gastric gavage, 200 mg/kg day) for the ICA group and sterandryl (subcutaneous injection, 5 mg/rat
day) for the ICA group and sterandryl (subcutaneous injection, 5 mg/rat day) for the T group for 7 consecutive days, respectively. The levels of serum testosterone, luteinizing hormone (LH), follicle stimulating hormone (FSH), serum bone Gla-protein (BGP) and tartrate-resistant acid phosphatase activity in serum (StrACP) were determined.
day) for the T group for 7 consecutive days, respectively. The levels of serum testosterone, luteinizing hormone (LH), follicle stimulating hormone (FSH), serum bone Gla-protein (BGP) and tartrate-resistant acid phosphatase activity in serum (StrACP) were determined.(1) Icariin improved increased the circulating levels of testosterone.
(2) Icariin treatment also improved the steady-state serum BGP and might have promoted bone formation. At the same time, it decreased the serum levels of StrACP and might have reduced the bone resorption.
Icariin has testosterone mimetic properties."
Icariin "stimulates the differentiation of osteoblasts and increases the activity of alkaline phosphatase (ALP) and the expression of I-collagen protein in rats and promotes bone formation"<-this does not mean that Icariin is anti-chondrogenic.
The increase in Testosterone by Icariin was insane, 3 fold versus the control group and only 1/3rd of the direct testosterone group.
Icariin promotes bone formation. BGP is also known as osteocalcin which is secreted by osteoblasts so it's used as a measure of bone formation. Therefore, Icariin increase bone formation. It's Icariin's usage as a testosterone mimetic that is intriguing however as myostatin inhibition has the most potential usage during endochondral ossification.
[Experimental study of icariin in inducing bone marrow mesenchymal stem cell differentiation]
"To research the effect of icariin (ICA) on the proliferation and differentiation of bone mesenchymal stem cells (BMSCs) and to study its influence on the expressions of transforming growth factor-beta1 (TGF-beta1) and bone morphogenetic protein-2 (BMP-2) in the progress of BMSCs differentiating into osteoblast, for providing an experimental evidence to explain the mechanism of ICA.
After the most effective concentration of ICA for promoting the differentiation of BMSCs into osteoblast was judged with the indices like alkaline phosphatase (ALP), etc., a grouped experiment was conducted for the sake of studying the effect and mechanism of ICA in its process of inducing BMSCs differentiation into osteoblast through detecting expression of ALP and calcium nodes, as well as the expressions of TGF-beta1, and BMP-2 with ELISA.
The most effective concentration of the ICA on the BMSCs differentiation was judged as 1 x 10(-9) mol/L, ICA of that concentration showed effects in enhancing the expressions of osteoblast-indices and increasing the secretion of TGF-beta1, and BMP-2. the increase of TGF-beta1, and BMP-2 was revealed in all the groups, in which ICA showed its influence visibly.
ICA could promote the differentiation of BMSCs into osteoblast; the up-regulation of TGF-beta1, and BMP-2 expressions is possibly one of the action mechanisms of various interventional drugs in their differentiation promoting progress."<-Note this shows that it increased BMP-2 secretion so definitely potential to induce chondrogenic differentiation. The study mentions encouraging the differentiation of MSCs into osteoblasts but both TGF-Beta1 and BMP-2 are pro-chondrogenic.
On the bone resorption inhibition(and remember bone resorption may be an important part of endochondral ossification):
Icariin inhibits osteoclast differentiation and bone resorption by suppression of MAPKs/NF-kappaB regulated HIF-1alpha and PGE(2) synthesis.
"We examined the detail molecular mechanisms of icariin on lipopolysaccharide (LPS)-induced osteolysis. Icariin [inhibits] osteoclast differentiation and bone resorption by suppressing MAPKs/NF-kappaB regulated HIF-1alpha and PGE(2) synthesis[HIF-1alpha and the NF-kappaB pathway are both good for chondrogenesis/height growth]. After treatment with icariin, the activity of osteoclasts differentiation maker, tatrate resistances acid phosphatease (TRAP), significantly decreased at the concentration of 10(-8)M. Icariin (10(-8)M) reduced the size of LPS-induced osteoclasts formation, and diminished their TRAP and acid phosphatease (ACP) activity without inhibition of cell viability. Icariin also inhibited LPS-induced bone resorption and interleukin-6 (IL-6)[at 10(-8)M], and tumor necrosis factor-alpha (TNF-alpha) expression. The gene expression of osteoprotegerin (OPG) was up-regulated, while receptor activator of NF-kappaB ligand (RANKL) was down-regulated. Icariin also inhibited the synthesis of cyclo-oxygenase type-2 (COX-2) and prostaglandin E(2) (PGE(2)). In addition, icariin had a dominant repression effect on LPS-induced hypoxia inducible factor-1alpha (HIF-1alpha) expression of osteoclasts. On osteoclasts, icariin suppresses LPS-mediated activation of the p38 and JNK; while on the osteoblasts, icariin reduced the LPS-induced activation of ERK1/2 and I-kappa-B-alpha (IkappaBalpha), but increased the activation of p38. Icariin has an in vitro inhibitory effects on osteoclasts differentiation that can prevent inflammatory bone loss. Icariin inhibited LPS-induced osteoclastogenesis program by suppressing activation of the p38 and JNK pathway."
"In macrophages, LPS induces hypoxia-inducible factor-1α (HIF-1α) protein accumulation under normoxic condition"<-this could be important for us as hypoxia is pro-chondrogenic.
Icariin has inhibitory effects on hypoxia in this study and chondrocytes like to grow in low oxygen environments but the inhibitory effects may be specific to LPS.
According to [Comparative study on effect of icariin and genistein on proliferation and mineralization of osteoblasts in vitro]., Icariin may also increase bFGF and IGF-1.
Icariin Promotes Extracellular Matrix Synthesis and Gene Expression of Chondrocytes In Vitro.
"Rabbit chondrocytes were isolated from articular cartilage and cultured in vitro with different concentrations of icariin. Icariin at concentrations under 1 × 10(-5) m showed low cytotoxicity toward chondrocytes, but icariin at 5 × 10(-5) m inhibited the proliferation of chondrocytes. Icariin hardly affected the cell morphology with concentrations ranging from 1 × 10(-7) m to 5 × 10(-5) m. the higher concentration of icariin produced more extracellular matrix (ECM) synthesis and expression of chondrogenesis genes of chondrocytes. Indeed, the promotion of icariin on the synthesis of glycosaminoglycans (GAGs) and collagen of chondrocytes, and finally exerting a potent chondrogenic effect, might be due to its ability to up-regulate the expression of aggrecan, collagen II and Sox9 genes and to down-regulate the expression of the collagen I gene of chondrocytes. Icariin [is] an effective accelerant for chondrogenesis. 1 × 10(-5) m may be a suitable concentration of icariin with chondrogenic effect for tissue engineering"
"icariin media at a concentration of 5 × 10−5 m inhibited the proliferation of chondrocytes, it violently promoted GAG synthesis of cells."<-Whether this is beneficial to height growth is the question. If chondrocytes are not proliferating they are not approaching senescence.
Icariin: Does It Have An Osteoinductive Potential for Bone Tissue Engineering?
"Icariin, a typical flavonol glycoside, is considered to be the main active ingredient of the Herba Epimedii from which icariin has been successfully extracted. Icariin can be delivered locally by biomaterials and that it has an osteoinductive potential for bone tissue engineering. The osteoinductive potential of icariin could be attributed to its multiple functions in the musculoskeletal system which is involved in the regulation of multiple signaling pathways in anti-osteoporosis, osteogenesis, anti-osteoclastogenesis, chondrogenesis, angiogenesis, and anti-inflammation."
This paper mentions a study where cell-hydrogel construct delivery of icariin in rabbits increased cartilage formation.
"Icariin inhibited LPS-induced osteoclastogenesis by suppressing the activation of the p38 and JNK pathway"
"The effect of icariin on the synthesis of glycosaminoglycans (GAGs) and collagen of chondrocytes, and its potent chondrogenic effect, might be due to its ability to up-regulate the expression of aggrecan, collagen II, and Sox9 genes and to down-regulate the expression of the collagen I gene of chondrocytes"
" Icariin can up-regulate the expression of Sox9 and Cbfa1 in controlling osteogenesis and chondrogenesis."
Labels:
dimethyl icariin,
horny goat weed
Subscribe to:
Posts (Atom)

