Recently, David stated that he had been loading his right epiphysis by 120lbs(!) to try to correct a 1cm length discrepancy. David used a hydraulic car jack and put two 45 lbs plates on his couch then he gradually used the jack to increase the pressure on his epiphysis. Here's his ankle picture after several months:
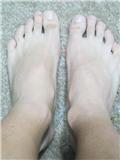
You can see his right epiphysis is much bigger than his left. David has reported no length change. An increase in bone size is excellent news because it indicates some of or some combination of the below: stem cell differentiation into osteoblasts which deposit new bone; mechanical signaling triggering existing osteoblasts to lay down new bone(bone modeling); and shear strain on periosteum resulting in periosteal progenitor cells which differentiate into osteoblasts which deposit new bone. Unfortunately, no height growth means no differentiation into chondroctyes. Could excessive loads favor osteogenesis over chondrogenesis? Or do you also need to load the articular cartilage to activate certain factors that favor chondrogenesis like Sox9 and TGF-Beta?
Signalling cascades in mechanotransduction: cell-matrix interactions and mechanical loading.
"Mechanical loading of articular cartilage stimulates the metabolism of resident chondrocytes and induces the synthesis of molecules to maintain the integrity of the cartilage[the articular cartilage connects to the hyaline cartilage of the growth plate so it can stimulate metabolism and synthesis of melcules there as well]. Mechanical signals modulate biochemical activity and changes in cell behavior through mechanotransduction. Compression of cartilage results in complex changes within the tissue including matrix and cell deformation, hydrostatic and osmotic pressure, fluid flow, altered matrix water content, ion concentration and fixed charge density[compression of cartilage changes hydrostatic pressure and one can presume growth plate hydrostatic pressure]. These changes are detected by mechanoreceptors on the cell surface, which include mechanosensitive ion channels and integrins that on activation initiate intracellular signalling cascades leading to tissue remodelling. Excessive mechanical loading also influences chondrocyte metabolism but unlike physiological stimulation leads to a quantitative imbalance between anabolic and catabolic activity resulting in depletion of matrix components[You shouldn't use too much load or risk depleting matrix components]. In this article we focus on the role of mechanical signalling in the maintenance of articular cartilage, and discuss how alterations in normal signalling can lead to pathology."
The bone begins as completely hyaline cartilage. Any mechanical loading induced growth should lead to increased long bone growth as the articular cartilage is connected to the resting zone hyaline cartilage until later in development.
"Increased joint loading in athletes is associated with an increase in the area of the load-bearing surface rather than an increase in cartilage thickness"<-Joint Loading increases joint width but not height but can lateral loading of joints encourage joint height as it's on a different axis?
"cyclic tensile loading increased the mRNA level of MMP-1, MMP-3, MMP-9, IL-1β, TNF-α and TIMP-1 in cultured chondrocytes"<-tensile loading means stretch. Aside from Interleukin Beta and TNF-Alpha these are the best genes for height growth to be expressed by chondrocytes. So lateral joint loading directly on the cartilage may have height increase benefits as well.
"insulin-like growth factor-1 (IGF-1) and TGF-β increase chondrocyte cell surface expression of α3/α5 integrin subunits and stimulate adhesion of chondrocytes to fibronectin and type II collagen"<-IGF-1 and TGF-Beta encourage chondrocytes to adhere to the Type II collagen parts of the growth plate
"IL-4 not only mediates anabolic signalling by increasing aggrecan expression but can decrease catabolic events. In an animal model intra-articular injection of IL-4 decreased chondrocyte nitric oxide production and inhibited destruction of cartilage in instability-induced osteoarthritis. Pre-treatment with IL-4 (10 ng/mL) suppressed both MMP-13 and cathepsin B induction by mechanical stress, as well as cyclical tensile stress-induced IL-1β expression"<-IL-4 may be a promising height increase supplement for the future.
Gene expression profiles of dynamically compressed single chondrocytes and chondrons.
"A chondrocyte produces a hydrated pericellular matrix (PCM); together they form a chondron. Previous work has shown that the presence of the PCM influences the biological response of chondrocytes to loading. The objective of this study was to determine the gene expression profiles of enzymatically isolated single chondrocytes and chondrons in response to dynamic compression. Cartilage specific extracellular matrix components and transcription factors were examined. Following dynamic compression, chondrocytes and chondrons showed variations in gene expression profiles. Aggrecan[increases proteoglycan content], Type II collagen[Type II collagen is the basis for articular and hyaline cartilage, could possibly lead to height growth] and osteopontin[involved in bone modeling] gene expression were significantly increased in chondrons. Lubricin gene expression decreased[Lubricin is a joint lubricant so could be part of a negative feedback mechanism] in both chondrons and chondrocytes. Dynamic compression had no effect on SOX9 gene expression. Our results demonstrate a clear role for the PCM in interfacing the mechanical signalling in chondrocytes in response to dynamic compression. Further investigation of single chondrocytes and chondrons from different zones within articular cartilage may further our understanding of cartilage mechanobiology."
Dynamic loading of articular cartilage may enhance height growth by upregulating Type II Collagen expression.
[Type II collagen fragment capacity to induce collagen cleavage and hypertrophy of articular chondrocyte]
"The objective of this study was to determine whether a peptide of type II collagen which can induce collagenase activity can also induce chondrocyte differentiation (hypertrophy) in articular cartilage. At high but naturally occurring concentrations (10 microM and up) the collagen peptide CB12-II induced an increase in the expressions of MMP-13 (24h) and cleavage of type II collagen by collagenase in the mid zone (day 4) and also in the superficial zone (day 6). Furthermore the peptide induced an increase in proliferation on day 1 in the mid and deep zones extending to the superficial zone by day 4. There was also upregulation of COL10A1 expression at day 4 and of type X staining in the mid zone extending to the superficial zone by day 6. Apoptopic cell death was increased by day 4 in the lower deep zone and also in the superficial zone at day 7. The increase in apoptosis in the deep zone was also seen in controls. Our results show that the induction of collagenase activity by cryptic peptide sequence of type II collagen is accompanied by chondrocyte hypertrophy and associated cellular and matrix changes. This induction occurs in the mid and superficial zones of previously healthy articular cartilage. This response of the chondrocyte to a cryptic sequence of denaturated type II collagen may play a role in naturally occurring hypertrophy in endochondral ossification and in the development of cartilage pathology in osteoarthritis."
So the Type II Collagen environment in the growth plate can trigger chondrocyte hypertrophy. This could be why proliferative capacity is conserved in growth hormone deficiency as there is less surrounding Type II Collagen. So overloading may result in the denaturing of Type II collagen which encourages ossification.
Genome-Wide Analyses of Gene Expression during Mouse Endochondral Ossification
"Numerous molecular markers characterize the central stages of the chondrocyte life cycle. Chondrogenesis is typified by the expression of Sox transcription factors 5,6 and 9. Proliferating chondrocytes synthesize an ECM composed mainly of collagen II and aggrecan, among others, while the central ECM molecule expressed in hypertrophic cartilage is collagen X. Factors expressed at the chondro-osseous junction regulate chondrocyte apoptosis and mineralization of the cartilaginous ECM. Late hypertrophic chondrocytes express factors that promote angiogenesis, bone deposition and the secretion of bone-specific cell ECM[So chondrocyte hypertrophy encourages ossification without estrogen]. These factors include Vegf (vascular endothelial growth factor), Mmp13(matrix metalloproteinase 13), Mmp9 and Ibsp. Additional markers of the osteoblast and osteoclast phenotype, including core-binding factor alpha 1/runt-related transcription factor 2 (Cbfa1/Runx2), acid phosphatase 5, tartrate resistant (Acp5) and tumor necrosis factor (ligand) superfamily, member 11 (Tnfsf11; RANKL/receptor activator of NF-kappaB ligand) are upregulated in hypertrophic cartilage and cells in the zone of ossification."
"When chondrocytes terminally differentiate, they undergo apoptosis, leaving behind a calcified extracellular matrix (ECM) that is remodeled and degraded by invading blood vessels, osteoprogenitor cells and bone-resorbing cells."<-terminal differentiation proceeds fusion which involves remodeling left over Extracellular Matrix. Ossification does not directly oppose cartilage growth.
"the expression of early stage chondrocyte markers such as Sox family members 5,6 and 9 (Sox5,6 and 9), Col2a1, growth differentiation factor 5 (Gdf5), Agc 1, Col11a1, Hapln1, Fgfr3 and Col9a2"<-These are the genes we are looking to be expressed by LSJL, not necessarily immediately but they should be expressed after LSJL is performed to indicate that LSJL was effective at inducing a new growth plate.
"several known growth plate markers [include] Sox9, Col2a1, Ihh and Cdkn1c"
Here's the list of genes upregulated by LSJL:
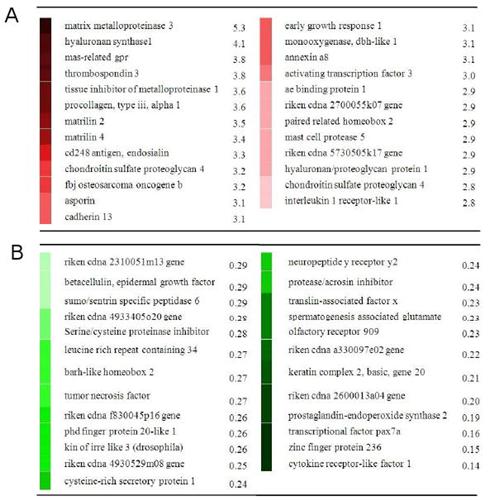
Column A are upregulated genes whereas Column B are downregulated. The genes involved are involved in the PI3K pathway and TGF-Beta pathway both of which are important to height growth. Perhaps, loading the cartilage is important to activate these genes and pathways. Note that the genes expressed shouldn't match the growth plate genes right away as it likely takes time for the growth plates to be formed. However, you should expect to see growth plate genes eventually.
Here's additional genes modified by LSJL:
Under expressed proteins are blue and overexpressed are red. There are no underexpressed proteins in the two pathways. When one point shares more proteins for example BMP's some proteins can be underexpressed and some over.
Other genes of note:
" Genes highlighted in those pathways include inositol 1,4,5-triphosphate 3-kinase and phospholipase C (plc) in PI3K, collagen (col3α1, col4α1, col6α1), integrin β4, and thrombospondin 3 in ECM-receptor interaction, TGFβ receptor 1 and smad1in TGFβ signaling pathway, and wnt2, frizzled 2 and Wnt1-inducible protein 2 in Wnt signaling pathway."
Single genes that can be identified as being overexpressed: Integrin Alpha V(CD51) and Integrin Alpha 11.
Genes that were upregulated above 2 fold or below .5 fold compared to control samples(Bolded genes are pro-chondrogenic):
Barx2
CDC42bpa
Zfp9
Cdh10
FGF2-2.433
ECM2-2.027
Dmrt2
Zfp533-2.188(upregulated during cartilage formation)
Gli3-3.184(one form of Gli3 may inhibit chondrocyte proliferation)
Fzd3
Nov-3.563
THBS2
Aspn
Dkk3
Itgbl1
Scx
H19-2.174(H19 expression is liked to IGF-2)
Grem2
FGFR1
Cav3-2.001(Myostatin inhibitor)
MMP2
THBS4
WISP2
SOCS3-2.484
TUBB6
Sox9-3.148
IGFBP6-2.083
Pax1-2.166
Agc1-2.474
Jun
Zfp36
Dnm1-2.352
Gas1
HMGA2-2.137(increased expression can cause overgrowth)
Dhh
COL2A1-2.012
MATN4-2.166
Zfp410
IRS1-0.339
PLRG1
TRAPPC3
TLR7
NPC1
Zfp148
Zfp106
HMGB2-0.291(reduction in HMGB2 is associated with apoptosis, according to Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation., HMGB2 inhibits chondrogenesis)
Zfp313
GPR108
Kif22
Zfp46
Rad17
Smad1-0.48
MAPK6
ADAMTS7-0.443
Zfp692
Zfp75
Zfp27
MAPK8
Zfp238
Id2-0.459(associated with early chondrogenic aggregates)
Gad1
GH-0.498
Lin9-0.471
STAT 5B-0.424(increases GH signaling)
Vcam 1-0.487(expressed by bone marrow stem cells, decrease in expression could indicate an increase in differentiation)
Accn1
MATN2-2.735
MATN3-2.485(Matrilin 3 is specific to cartilage matrix proteins)
Nkx3.2(also known as Bpax1)-0.49(this gene is pro-chondrogenic so it may be downregulated as part of a negative feedback mechanism)
Wnt2-3.089
Hey2-2.685
ADAMTS1-2.674
Hes1-2.405
Smad9-2.383
dusp14-2.32
Arg1-2.25
Sulf1-2.195
Vcan-2.144
Lin28B-2.143(Increases HMGA2 activity which in turn should increase height)
MMP14-2.055
HHIP-2.014
MMP7-0.491
CAMK2G-0.458
TGFBR1-0.411
Arg2-0.36
HTR2C-10.47(Serotonin Receptor)
PTGS2-7.63(synthesizes prostoglandin)
TNMD-6.185(Tendon Molecular marker)
IL6-3.051(activates STAT3)
BMPR1B- 2.166
COL9A1-2.455
COL11A1-2.029
HAPLN1-2.585(expressed in early chondrogenesis, associated with hyaluronic acid binding)
Dpt-3.144(increases in expression throughout chondrogenesis)
PRKG2-2.894(associated with cGMP)
Ptn-2.649(regulates cell cycle)
PDGFC-2.1
PKIA-2.013(inhibits cAMP)
CCNB1-0.437(cell cycle progression)
Sdc2-0.426(Syndecan 2, involved in pre-chondrogenic differentiation)
PDE6H-0.424(associated with cAMP)
HABP4-0.417(binds with hyaluronic acid)
CREB3L1-2.238(also called OASIS, may increase GH and IGF-1 levels)
COL10A1-2.012
HTRA1-2.322(interacts with BMP4, Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. states that HtrA1 is involved in Aggrecan breakdown)
HES5-2.562(In the data as BHLHB5)
GNAS-0.329(implicated in some forms of heterotropic ossification)
According to another LSJL study LSJL suppresses Sost and Dkk1.
Here's how these genes compare to MSCs normally undergoing endochondral ossification.
Gene Expression Profile during Chondrogenesis in Human Bone Marrow derived Mesenchymal Stem Cells using a cDNA Microarray.
"Chondrogenesis was induced by culturing human bone marrow (BM) derived MSCs in micromass pellets in the presence of defined medium for 3, 7, 14 or 21 days. Several genes regulated during chondrogenesis were then identified by reverse transcriptase-polymerase chain reaction (RT-PCR). Using an ABI microarray system, we determined the differential gene expression profiles of differentiated chondrocytes and BM-MSCs. Normalization of this data resulted in the identification of 1,486 differentially expressed genes. To verify gene expression profiles determined by microarray analysis, the expression levels of 10 genes with high fold changes were confirmed by RT-PCR. Gene expression patterns of 9 genes (Hrad6B, annexinA2, BMP-7, contactin-1, peroxiredoxin-1, heat shock transcription factor-2, synaptotagmin IV, serotonin receptor-7, Axl) in RT-PCR were similar to the microarray gene expression patterns."
Note that the LSJL gene expression study was done on rats whereas this was done on humans. It's also possible that these genes were expressed at lower levels than were noted in the study. Note there is definite signs of chondrogenesis like the genes related to proteoglycans.
These are different than the highly expressed genes in the LSJL study, AnnexinA2 facilitates actin cytoskeleton formation whereas in the LSJL study AnnexinA8 was present. AnnexinA8 is an anticoagulant so it stops blood clotting likely to reduce hydrostatic pressure.
"The antigen which bound to SH2 antibody was identified as endoglin (CD105), a receptor for TGF-β3, which potentially plays a role in mediating the chondrogenic differentiation of MSCs and in their interactions with hematopoietic cells"<-endosialin is present instead in LSJL. Endosialin relates to calcium ion binding.
"Axl, synaptotagmin IV, Hrad6B, peroxiredoxin-1, BMP-7, heat shock transcription factor-2, annexin A2, contactin-1 and serotonin receptor-7 expressions were maintained in differentiating BM-MSCs until day 14."<-So maybe upregulating serotonin expression can upregulate chondrogenesis?
Wnt2-3.089
Hey2-2.685
ADAMTS1-2.674
Hes1-2.405
Smad9-2.383
dusp14-2.32
Arg1-2.25
Sulf1-2.195
Vcan-2.144
Lin28B-2.143(Increases HMGA2 activity which in turn should increase height)
MMP14-2.055
HHIP-2.014
MMP7-0.491
CAMK2G-0.458
TGFBR1-0.411
Arg2-0.36
HTR2C-10.47(Serotonin Receptor)
PTGS2-7.63(synthesizes prostoglandin)
TNMD-6.185(Tendon Molecular marker)
IL6-3.051(activates STAT3)
BMPR1B- 2.166
COL9A1-2.455
COL11A1-2.029
HAPLN1-2.585(expressed in early chondrogenesis, associated with hyaluronic acid binding)
Dpt-3.144(increases in expression throughout chondrogenesis)
PRKG2-2.894(associated with cGMP)
Ptn-2.649(regulates cell cycle)
PDGFC-2.1
PKIA-2.013(inhibits cAMP)
CCNB1-0.437(cell cycle progression)
Sdc2-0.426(Syndecan 2, involved in pre-chondrogenic differentiation)
PDE6H-0.424(associated with cAMP)
HABP4-0.417(binds with hyaluronic acid)
CREB3L1-2.238(also called OASIS, may increase GH and IGF-1 levels)
COL10A1-2.012
HTRA1-2.322(interacts with BMP4, Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. states that HtrA1 is involved in Aggrecan breakdown)
HES5-2.562(In the data as BHLHB5)
GNAS-0.329(implicated in some forms of heterotropic ossification)
According to another LSJL study LSJL suppresses Sost and Dkk1.
Here's how these genes compare to MSCs normally undergoing endochondral ossification.
Gene Expression Profile during Chondrogenesis in Human Bone Marrow derived Mesenchymal Stem Cells using a cDNA Microarray.
"Chondrogenesis was induced by culturing human bone marrow (BM) derived MSCs in micromass pellets in the presence of defined medium for 3, 7, 14 or 21 days. Several genes regulated during chondrogenesis were then identified by reverse transcriptase-polymerase chain reaction (RT-PCR). Using an ABI microarray system, we determined the differential gene expression profiles of differentiated chondrocytes and BM-MSCs. Normalization of this data resulted in the identification of 1,486 differentially expressed genes. To verify gene expression profiles determined by microarray analysis, the expression levels of 10 genes with high fold changes were confirmed by RT-PCR. Gene expression patterns of 9 genes (Hrad6B, annexinA2, BMP-7, contactin-1, peroxiredoxin-1, heat shock transcription factor-2, synaptotagmin IV, serotonin receptor-7, Axl) in RT-PCR were similar to the microarray gene expression patterns."
Note that the LSJL gene expression study was done on rats whereas this was done on humans. It's also possible that these genes were expressed at lower levels than were noted in the study. Note there is definite signs of chondrogenesis like the genes related to proteoglycans.
These are different than the highly expressed genes in the LSJL study, AnnexinA2 facilitates actin cytoskeleton formation whereas in the LSJL study AnnexinA8 was present. AnnexinA8 is an anticoagulant so it stops blood clotting likely to reduce hydrostatic pressure.
"Axl, synaptotagmin IV, Hrad6B, peroxiredoxin-1, BMP-7, heat shock transcription factor-2, annexin A2, contactin-1 and serotonin receptor-7 expressions were maintained in differentiating BM-MSCs until day 14."<-So maybe upregulating serotonin expression can upregulate chondrogenesis?
"BMP-7 is a strong chemotactic component in cartilage cells produced by mesenchymal stem cells, and it can promote cartilage cells to secrete specific extracellular matrix (proteoglycans and collagen type II). And BMP-7 can induce the differentiation of BM-MSCs into cartilage cells."
"Annexins bind to negatively charged phospholipids in a Ca2+-dependent manner."
"Contactin-1 is a cell surface adhesion molecule"
"IL-15 [which is expressed by MSCs] is a potent apoptosis inhibitor "
Hiroki Yokota wrote a paper that mentions gene expression in chondrogenesis:
Modelling and identification of transcription-factor binding motifs in human chondrogenesis.
Hiroki Yokota wrote a paper that mentions gene expression in chondrogenesis:
Modelling and identification of transcription-factor binding motifs in human chondrogenesis.
Red are downregulated genes and green are upregulated genes. Black is neutral. We're looking for genes upregulated 1 day as that would be the time frame closest to that of the LSJL gene expression study. LSJL genes were taken at 49 hours. Column A is the observed profile. ILR1(although in LSJL it's interleukin 1 receptor-like 1) and BMP2 is shared between chondrogenesis and LSJL.
The predicted regulatory model for Col2A1 "[predicts] that AP-1 and Smad would be the continuous stimulator, and Sox9 would be the inhibitor at day 1 and the stimulator at days 7 to 21". c-Fos which LSJL upregulates is part of the AP-1 complex.
"AP-1 is reported to play the critical role in differentiation as a target of chondrogenetic growth factor such as bone morphogenetic protein-2 (BMP-2)"
"The model predicted that AP-1 would be the strong stimulatory factor during chondrogenesis. NFkB is known to control expression of BMP-2 and Sox-9 genes"
"The Sox-9 gene is one of the essential transcription factors in chondrogenesis by activating the enhancer element of a series of chondrogenetic marker genes such as Col2a1; Col9a2; Col11a2; and Aggrecan"


This blog entry mentions weight applied but neglects to mention the length of time per loading session, times per day, and days per week over the several month period. That's IMPORTANT to note.
ReplyDeleteAlso, does David post on the forum here? If so, by what name?
So, Tyler are you suggesting what we shouldn't clamp too hard or overload the epiphysis cause it depletes the matrix components, is that right?!
ReplyDeleteI clamp everyday for 3 minutes at medium intensity, is it excessive?
Well, I just proved that LSJL upregulates chondrogenesis at the intensity used in the study. It's possible that too much intensity is detrimental to height growth but other evidence indicates otherwise.
DeleteRight now what I'm doing is 4 days loading legs and then 4 days loading arms to get around the conditioning effect.
Right I'm loading for four minutes medium intensity at that loading regime.